Developed One-step Reverse Transcription Loop-Mediated Isothermal Amplification (Rt-Lamp) Assay for Detection of Strawberry Latent Ring Spot Virus
Developed One-step Reverse Transcription Loop-Mediated Isothermal Amplification (Rt-Lamp) Assay for Detection of Strawberry Latent Ring Spot Virus
Ahmed A. Kheder
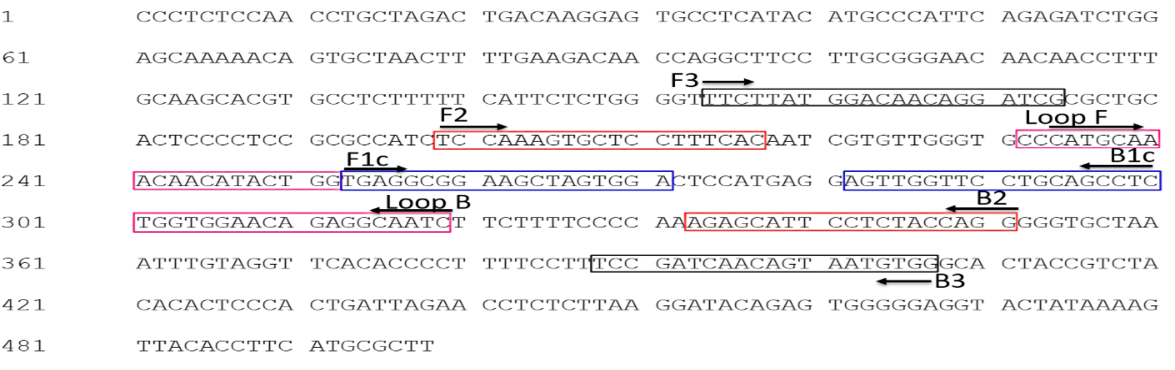
Targeted amplification region on the coat protein gene RNA2 of Strawberry latent ring spot virus (MT648777.1) for RT-LAMP assay.
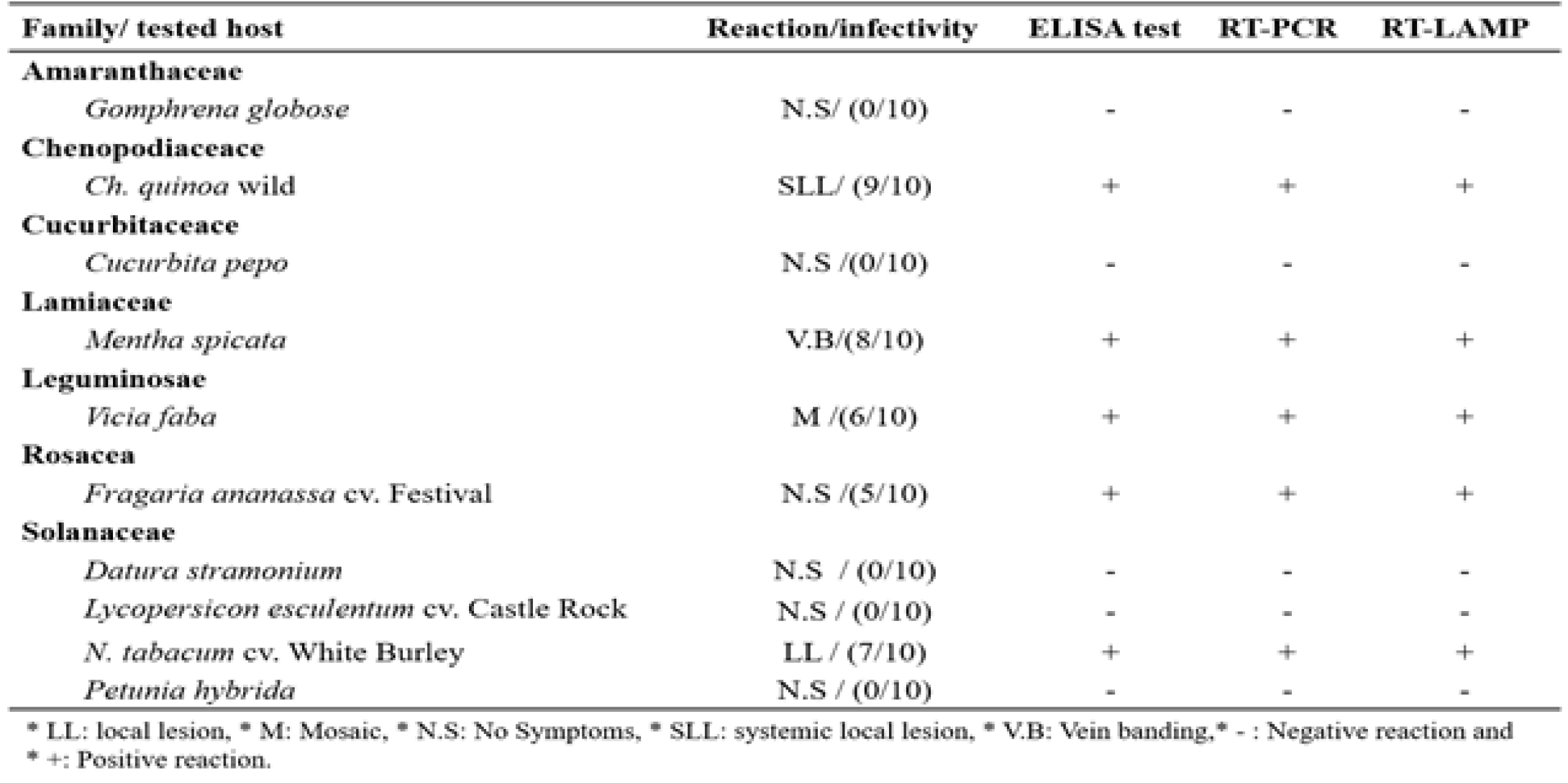
Symptoms of Strawberry latent ring spot virus (SLRSV). Naturally infected strawberry plants showing viral like symptoms (A) symptomless plant (B) showing mottling. Mechanically inoculated herbaceous hosts: (C and D) mechanically inoculated Fragaria ananassa cv. Festival leaves showed yellowing and mottling, (E) systemic local lesion on Chenopodium quinoa, (F) Chlorotic local lesion on Nicotiana tabacum cv. White Burley, (G) mosaic vein clearing and chlorosis on Vicia faba plant and (H) vein banding on Mentha spicata.
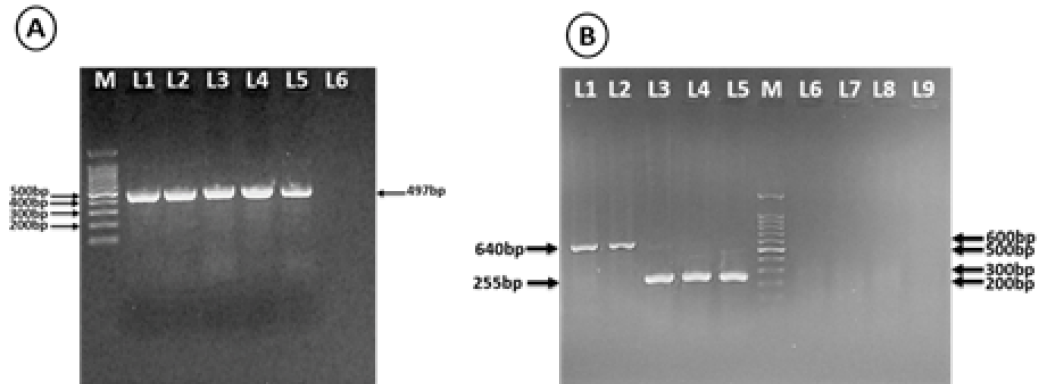
Agarose gel electrophoresis analysis of one-step RT-PCR amplified products of naturally infected and mechanically inoculated plants. (A) Using CP-RNA2 specific primer, Lane 1: Naturally infected strawberry plants, Lane 2: inoculated Ch. quinoa wild, Lane 3: inoculated Vicia faba, Lane 4: inoculated Fragaria ananassa cv. Festival, Lane 5: inoculated N. tabacum cv. White Burley, Lane 6: empty and M: 100pb Marker (Biomatic-USA). (B) using nepovirus degenerate primers, Lane1: ToRSV as a positive control of group C, Lane 2: naturally infected strawberry plant detection with Nepo-C (s/a) Lane 3: GFLV as a positive control of group A, Lane 4-5: Naturally infected strawberry plant detection with Nepo-A (s/a), Lane 6, 7 and 8: Naturally infected strawberry plant with SLRSV detected with Nepo-A (s/a), Nepo-B (s/a) and Nepo-C (s/a) respectively, Lane 9: Negative control and M: 100pb Marker (Biomatic-USA).
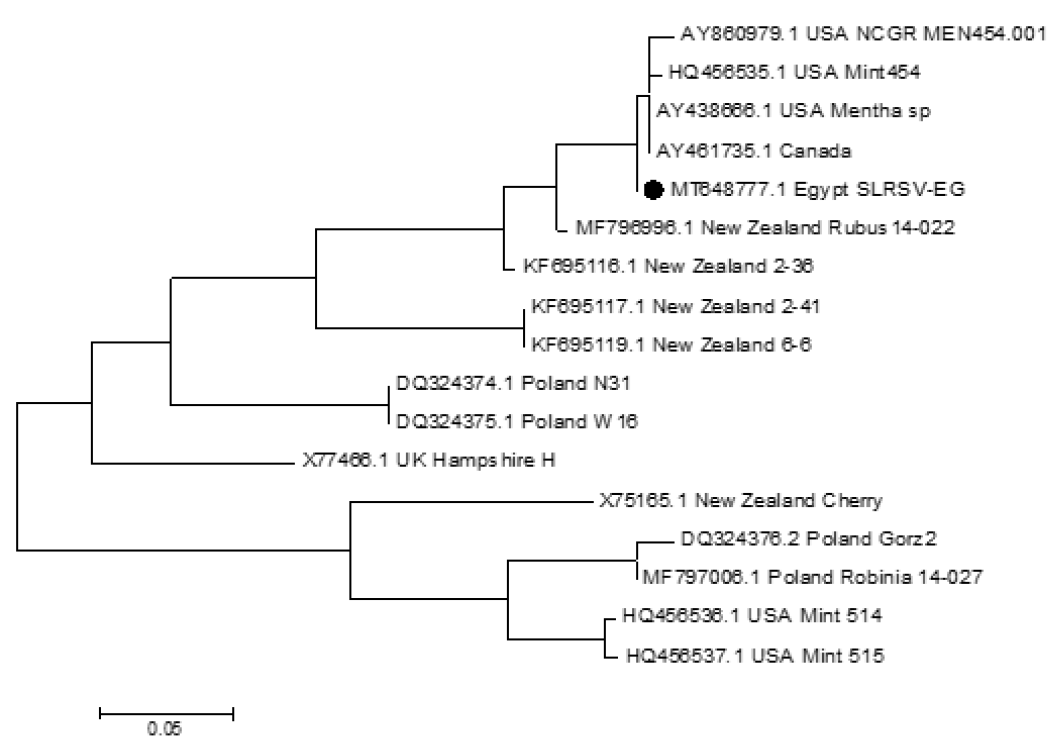
Phylogenetic relation of Strawberry latent ring spot virus isolates based on a fragment of RNA2. The numbers at each branch point indicate bootstrap values, branch values shown are also 99.5 – 65.9%. Nucleotide sequences used for above analysis available in GenBank.
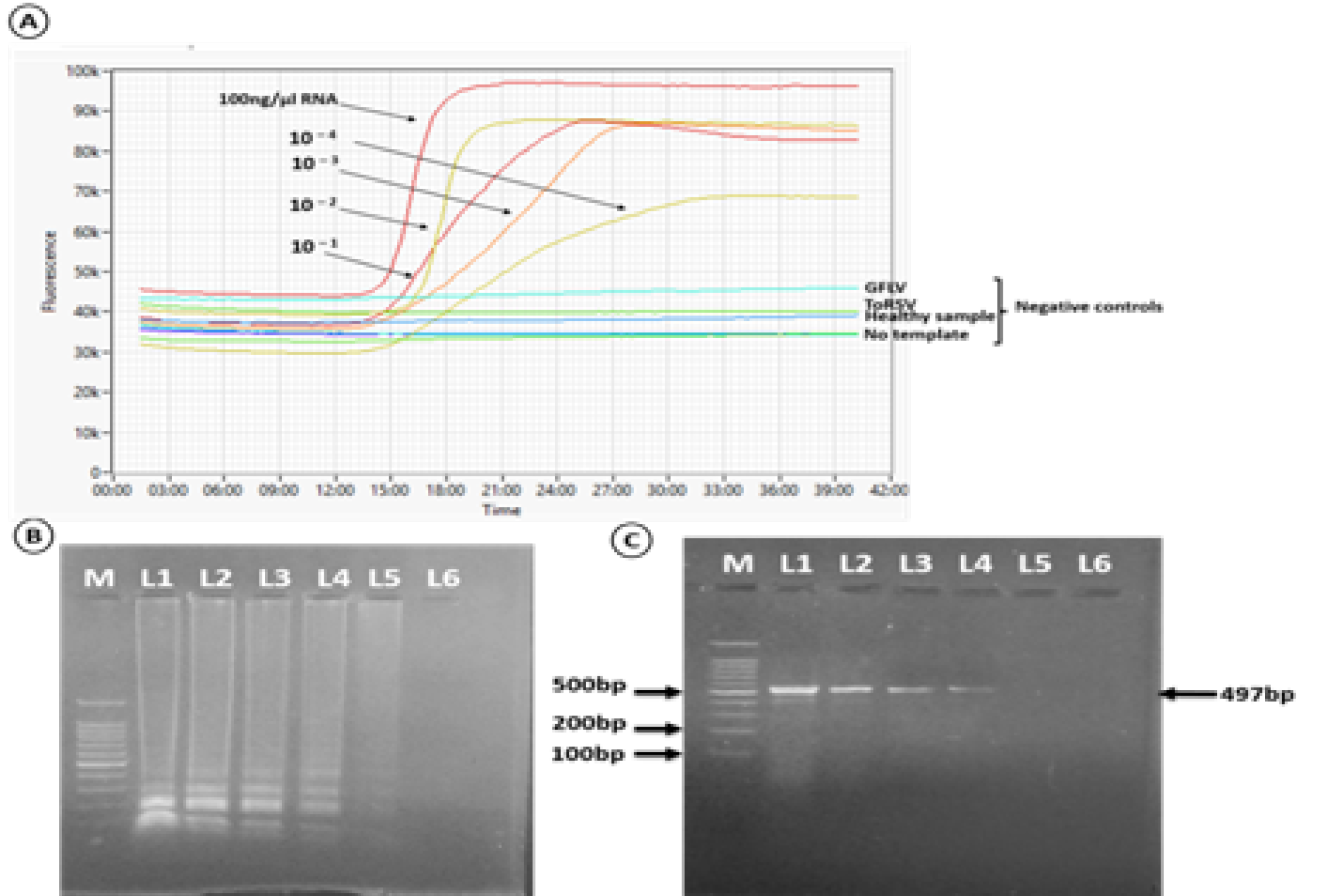
Sensitivity comparison between the RT-LAMP assay and the RT-PCR detection method. The detection limit of the RT-LAMP assay was determined based on real-time amplification plots and no visible amplification of GFLV, ToRSV and healthy samples as well as water control (A), Gel electrophoresis (b) and one-step RT-PCR (C). M. 100pb molecular marker (Biomatic-USA); Lane 1, total RNA 100ng/µl; Lane 2-6 tenfold serial dilutions of total RNA. The order of samples in panel C is the same as in panel B.
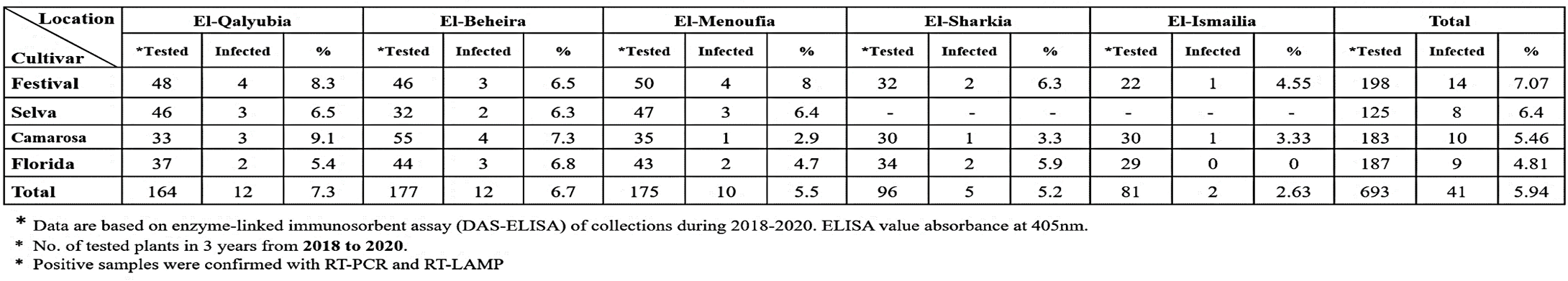
Incidence of Strawberry latent ringspot virus (SLRSV) infection in strawberry plants in different location in Egypt.
Reaction of some inoculated host plants to SLRSV infection.