In Silico Analyses of the Pseudogenes of Helicobacter pylori
In Silico Analyses of the Pseudogenes of Helicobacter pylori
Neenish Rana, Nosheen Ehsan, Awais Ihsan and Farrukh Jamil*
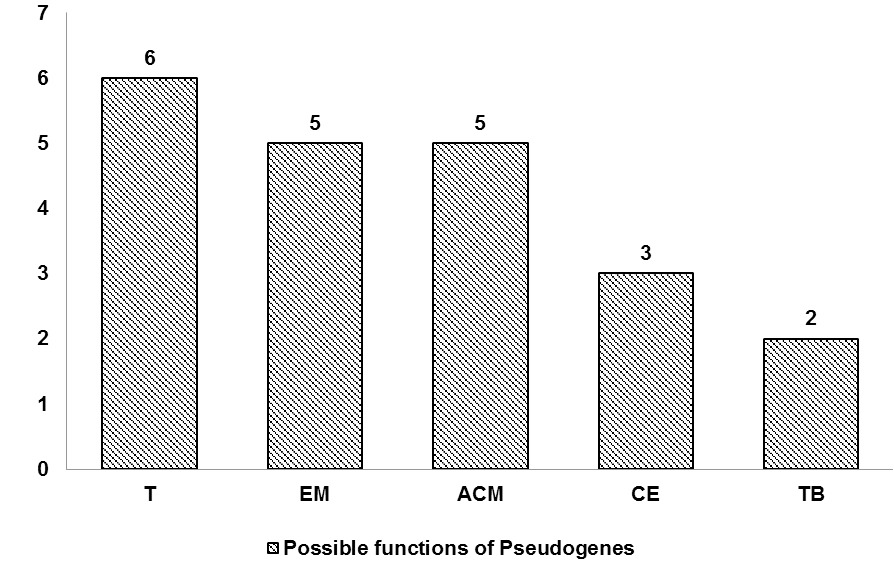
Function prediction of pseudogene-derived proteins. 6 proteins were predicted to be involved in translation (T), 5 in energy metabolism (EM), 5 in amino acid metabolism (ACM), 3 in cell envelope (CE) while, the functions of 2 proteins predicted in transport and binding (TB).
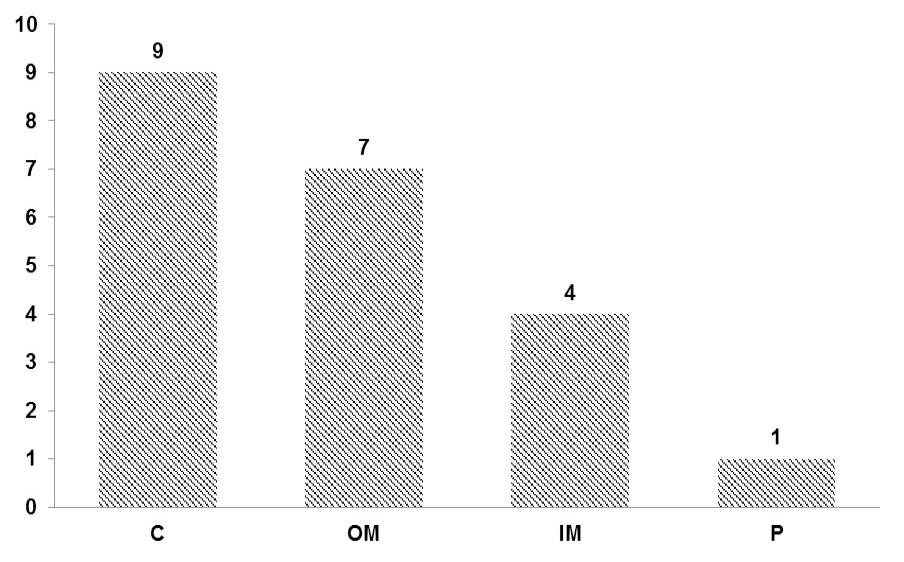
Sub-cellular localization of pseudogene-derived proteins. 9 proteins showed high potential to be localize in cytoplasm (C), 7 proteins appear to reside in outer membrane (OM), 4 proteins localized to inner membrane (IM) while localization of remaining 1 predicted in periplasm (P).