Refolding of Misfolded Inclusion Bodies of Recombinant α-Amylase: Characterization of Cobalt Activated Thermostable α-Amylase from Geobacillus SBS-4S
Refolding of Misfolded Inclusion Bodies of Recombinant α-Amylase: Characterization of Cobalt Activated Thermostable α-Amylase from Geobacillus SBS-4S
Sabah Mansoor1, Muhammad Tayyab1,*, Amna Jawad1, Bushra Munir2, Sehrish Firyal1, Ali Raza Awan1, Naeem Rashid3 and Muhammad Wasim1
Phylogenetic tree: The tree was constructed using amino acid sequence of amylase from Geobacillus SBS-4S and reported sequences from NCBI GenBank. The name at the end of each branch present the bacterial strain with accession number from which the amylase sequence was originated. Clades A, B and C present the GH family 13, 77 and 70, respectively. The tree was constructed at a genetic distance of 0.5 using Mega 4 software.
Amino acid sequence comparison of AMYSBS (AB971162) with its closest homologue G. thermoleovorans (4E2O) that has been characterized. Identical amino acids are shown by asterisks below the sequence. The names at left hand side, indicates the organism from which the sequence originated. The active site residues are shown by bold letters. The open and closed circles above the sequence represent the amino acids involved in the binding of metal ions.
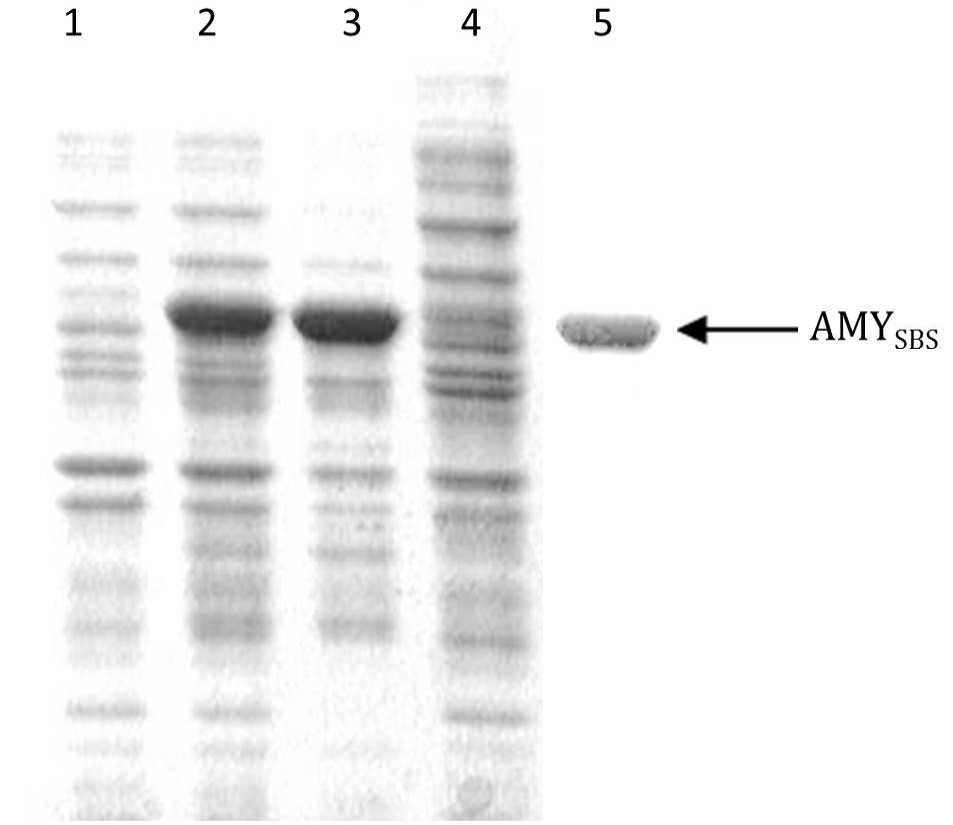
Coomassie brilliant blue stained sodium dodecyl sulphate polyacrylamide gel showing expression of AMYSBS gene: Lane 1, the soluble portion after lysis of BL21-CodonPlus (DE3) cells transformed with pET-21a without insert (negative control); Lane 2, the total cell protein after lysis of BL21-CodonPlus (DE3) cells transformed with pET-AMY; Lane 3, insoluble portion after lysis of sample in lane 2; Lane 4, soluble portion after lysis of sample in lane 2; Lane 5, purified AMYSBS after column chromatography.
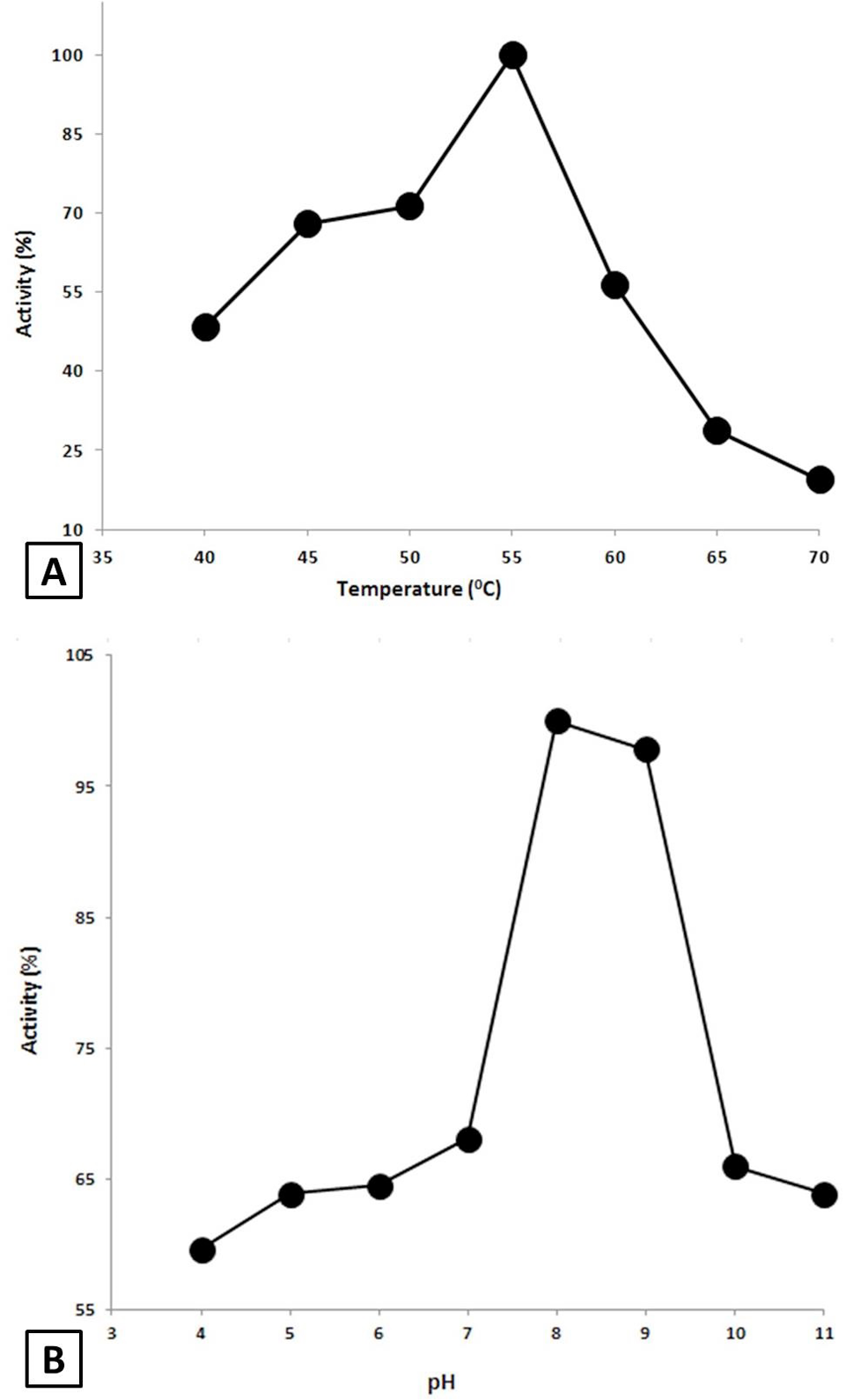
Effect of Temperature and pH on the AMYSBS activity. A, Effect of temperature. The activity was done at various temperatures ranging from 40 to 70°C in 50 mM Tris-HCl buffer (pH 8) using 0.4% starch as substrate. B, Effect of pH on the activity of AMYSBS. The activity was examined in 50 mM of each of acetate buffer (4-5), phosphate buffer (6-7), Tris-HCl buffer (8-9) and Glycine/NaOH (10-11) using 0.4% starch as substrate at 55°C.
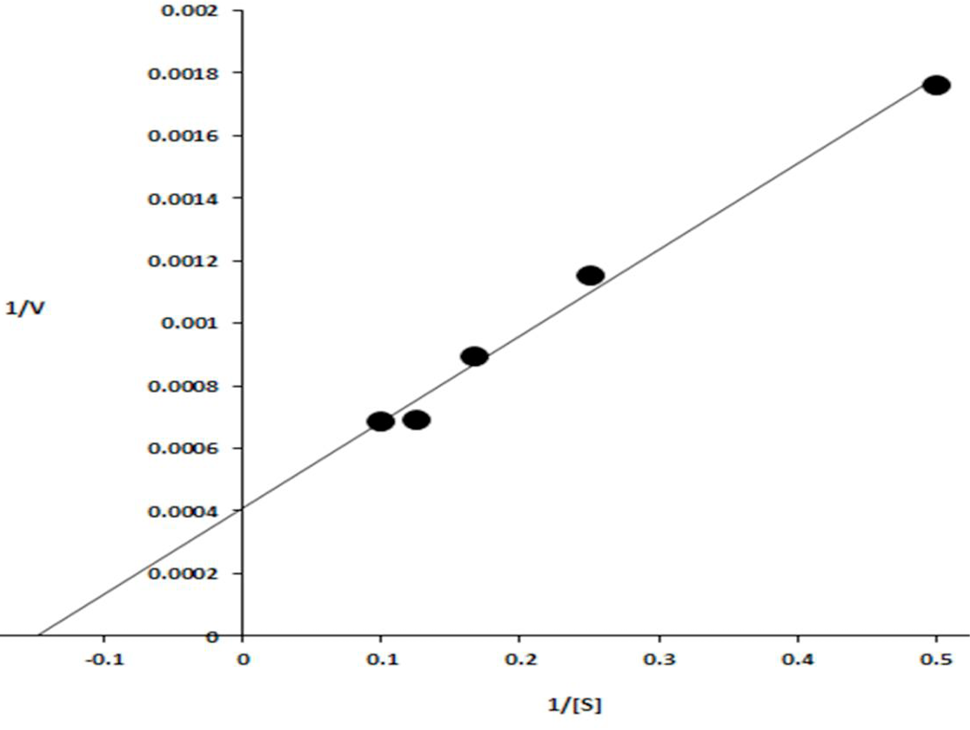
Lineweaver–Burk plot obtained by taking the inverse of the substrate concentrations (mg/ml) along X-axis and velocities (μmol min−1 mg-1) along Y-axis.