Characterization and Bio-Antagonistic Activity of Rhizobacteria against Fusarium Oxysporum F. Sp. Cepae
Characterization and Bio-Antagonistic Activity of Rhizobacteria against Fusarium Oxysporum F. Sp. Cepae
Javed Asghar Tariq1*, Bashir Ahmed2, Manzoor Ali Abro2, Muhammad Ismail1, Muhammad Usman Asif1 and Raza Muhammad1
1Nuclear Institute of Agriculture Tando Jam, Pakistan; 2Deptartment of Plant Pathology, Sindh Agriculture University, Tando Jam, Pakistan.
Abstract | Rhizobacteria have ability to enhance plant growth as well as to have antagonistic effect against another pathogenic microflora. Current studies were designed with the aim to search out such type of antagonistic bacteria from rice crop. For this purpose, soil was collected from rice growing areas of Tando Jam. The isolation from rhizospheric soil was done by dilution plate technique. Among isolated isolates, 6 representative colonies were purified, multiplied and characterized using light microscopy and other biochemical tests. The bacteria were tentatively identified as Pseudomonas, Streptomyces, Bacillus and Micrococus. These bacteria were checked for their antagonistic activity against Fusarium oxysporum f.sp. alium cepa. They all were found to inhibit fungus showing inhibition zone 4.09 -74.97%. Hence, on the basis of these studies, it can be concluded that these bacteria can be used as a biocontrol agent to manage other fungal diseases of agricultural crops also.
Received | March 20, 2018; Accepted | Febraury 16, 2019; Published | April 25, 2019
*Correspondence | Javed Asghar Tariq, Nuclear Institute of Agriculture Tando Jam, Pakistan; Email: jatariq_1411@yahoo.com
Citation | Tariq, J.A., B. Ahmed, M.A. Abro, M. Ismail, M.U. Asif and R. Muhammad. 2019. Characterization and bio-antagonistic activity of rhizobacteria against Fusarium oxysporum F. Sp. Cepae. Pakistan Journal of Agricultural Research, 32(2): 353-358.
DOI | http://dx.doi.org/10.17582/journal.pjar/2019/32.2.353.358
Keywords | Rhizobacteria, Biocontrol, Diseases, Fungi, Crops
Introduction
Soil is an excellent medium for the growth of several microorganisms including viruses, fungi, protozoa and bacteria. Microorganisms like rhizobacteria have ability to colonize rhizosphere (the plant roots), making a beneficial association with tertiary roots and root hairs (Kennedy, 2005). As a result of this association, these bacteria increase their population and adhere to plant roots during all stages of plant growth. Generally, association between micro-organisms and plants can be divided as beneficial, pathogenic and saprophytic (Lynch, 1990). Beneficial association involves adhering or sticking of plant growth promoting rhizobacteria (PGPR) with roots in a competitive soil medium and employing a useful effect on the plant improvement (Kloepper and Schroth, 1978; Lazarovits and Nowak, 1997; Kloepper, 1989; Bakker et al., 2007). Over past several years, PGPR have been renowned as beneficial plant microbes, being helpful in increasing plant growth and enhancing crop production. Currently these rhizobacteria are used repeatedly in field experiments by several researchers. However, in different studies by Farzana et al. (2009) and Munase and Mulugeta (2014) it has been reported that such bacteria improved the yield of sugar beet, potato, sweet potato and radish successfully. The bacterial species belonging to genera like Azospirillum, Bacillus, Alcaligenes, Serratia, Pseudomonas, Arthrobacter, Enterobacter, Acinetobacter, Burkholderia, Flavobacterium, Erwinia, and Rhizobium, are also related with which the plant rhizosphere having beneficial influence on plant growth (Tilak et al., 2005). It has also been described that PGPR can be used by replacing the different chemicals in the shape of pesticides, fertilizers and other supplements (Zaman et al., 2009). Furthermore, it has been been observed that approximately 10 to 20 percent loss in production is caused by plant diseases (James, 1981; Serge et al., 2012) which needs an attentive management strategy. The use of antibacterial and antifungal chemicals is criticized due to their harmful effects. So, a substitute to chemicals is the use of bacteria to control plant diseases which is being considered as a more environmentally friendly process (Van Loon and Glick, 2004; Merina et al., 2015). Rhizobacteria can suppress other microorganisms by secreting their metabolites i.e. antibiotics and lytic enzymes and through competition (Van Loon and Bakker, 2003) that make them a potent tool for reducing damages through preventing deleterious effects of other phytopathogens. Current studies were designed with the aim to search out such type of bacteria from rice rhizosphere, which can enhance plant growth as well as save the plants from deleterious soil micro-organisms.
Materials and Methods
Collection of soil samples
Collection of soil samples was done from different areas of Tando Jam from rice rhizosphere for isolation, purification and characterization of rhizobacteria.
Isolation of Rhizobacteria
Isolation of rhizobacteria was done by serial dilution. For this purpose, 200 grams of soil from rice field was processed for isolation of rhizobacteria. One gram of soil was diluted in distilled water in conical flask and then this mixture was vortexed and dilutions were made up to 10-8 using glass test tubes. From each dilution, 0.1ml was poured on already prepared Nutrient Agar media (N.A) plates. Then these poured petri plates were placed at 28 ± 2 0C for 3 days for incubation.
Purification of Rhizobactrial cultures
A total of six selected rhizobacterial isolates were purified by streaking method which involves separating and spreading of rhizobacteria on nutrient agar plates with the help of inoculating loop.
Characterization and identification of Rhizobacterial isolates
Rhizobacteria were characterized and identified on morphological as well as biochemical basis i.e. cell morphology, colony morphology, gram staining, catalase and amylase activity.
Gram staining
On the basis of physical and chemical composition, Gram staining method was used for differentiating bacterial species as Gram positive or Gram-negative. The smear was prepared from 1-2 drops of culture on clean slide and heat fixed. 1-2 drops of crystal violet solution were applied on the fixed smear for 1 min and then washed with sterile distilled water. Gram’s iodine solution was applied for 1 min and then washed with 95% alcohol. Finally, the smear was stained with counter stain Safranin for 30 seconds, and again washed with sterile distilled water. The smears were air dried and examined under light microscope by using oil immersion. The Gram positive bacterial cells appeared violet while gram negative bacteria turned pink to red (Vincent, 1947). Again, the slide was washed and blot dried. The slide was observed under microscope using immersion oil.
Catalase Test: Catalase test was done as described by Joint N.Q. (2016) by placing a drop of hydrogen peroxide on a microscope slide having bacterial smear. The bacterial smear producing bubbles or froth were said to be ‘catalase-positive.’ If the mixture did not produce bubbles or froth, the organism was said to be ‘catalase-negative’.
Amylase Test: (Starch Hydrolysis): Starch hydrolysis test were performed to determine the ability of rhizobacteria to use starch as a carbon source (De Oliverira, 2007). The medium was inoculated with rhizobacteria and analyzed for starch utilization. Iodine test was used to determine the capability of rhizobacteria to use starch. Drops of iodine solution (0.1 N) were spread on 24 hours old cultures grown in petri plates. A color change i.e. formation of blue color were indicated non utilization of starch and vice versa.
Salt, pH and temperature tolerance
The ability of rhizobacterial strains to grow at different concentration of salt was tested by the method as described by Suresh at el. (2013). It was done by streaking bacteria on NA medium containing 1.0%, 3.0% and 5% salt, (wt/v) i.e. NaCl. Differences in pH
Table 1: Morphological characterization of rhizobacterial isolates.
|
Train No.
|
Colony Morphology | Cell Morphology | Tentative Identification | |||||||
| Size | shape | Elevation | Edges | Color | Surface | Shape | Mo-tile Y/N | Gram rea- ction |
||
| BRS 15 | Medium | Filamen-tous | Raised | Undulate | Off White | Smooth | Selender | Y | + | Streptomyces |
| BRS 24 | Medium | Irregular | Convex | Filamentus | Off White | Smooth | Rod | N | + | Bacillus |
| BRS- 3 | Large | Spindle | Convex | Undulate | Yello wish |
Smooth | Cocci | N | + | Micrococus |
| BRS- 4 | Small | Circular | Convex | Entire | Red | Smooth | Rod | N | + | Streptomyces |
| BRS- 9 | Large | Spindle | Convex | Undulate | Yello wish |
Smooth | Cocci | N | - | Pseudomonas |
| BRS- 6 | Small | Circular | Convex | Undulate | Pale yellow | Smooth | Rods | N | - | Pseudomonas |
tolerance were tested by adjusting the pH to 6.5, 7.5, and 8.0. Difference in the range of growth temperature was investigated by incubation of rhizobacterial cultures at 35°C, 40°C and 45°C. Control plates were incubated at 28°C. Strains were considered salt tolerant, resistant to pH and temperature resistant when growth was found to be similar to the growth in the control plates.
Statistical analysis
The data obtained by these studies was subjected to analysis of variance (ANOVA) for a completely randomized design and the means were compared using post-hoc Turkey’s HSD test with P< 0.05 being accepted as significance.
Antagonistic test between rhizobacteria and Fusarium oysporum f. sp. cepae antifungal activity
The antifungal activity of rhizobacterial strains against Fusarium oxysporum f. sp. cepae was checked on PDA medium (Potato Dextrose Agar) by dual culture method as described by Gupta et al. (2001). An agar bit (5 mm diameter) of 5-day-old culture of test fungus was placed in the center of PDA plates. A loopful of 24-h-old culture of rhizobacterial strain was then streaked at either side of fungal bit at a distance of 2 cm and un-inoculated plates (by rhizobacterial strain), was served as control. The treatments were replicated thrice and were incubated at 25±1ºC for 5 days. Percentage inhibition produced by the rhizobacterial strain against Fusarium oysporum f. sp. cepae and the control was calculated by using the formula given by Vincent (1947) as follows:
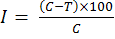
Where;
I= Percent inhibition of fungal mycelia; C= Growth of mycelium in NA plates (served as the control); T= Growth of mycelium in the treatment.
Results and Discussion
Isolation, purification and morphological characterization of rhizobacteria isolated from rice crop
Several rhizobacterial colonies appeared on NA medium after incubation at 280C. Among them six purified colonies were selected for further studies. Isolated and purified rhizobacterial strains were tentatively identified as genus Micrococus, Streptomyces, Pseudomonas and Bascillus. The colonies of isolated rhizobacteria on nutrient agar (NA) were circular, convex, off white, smooth and rod. Streptomyces were to be gram positive and motile. Their colonies on NA media were irregular, convex, filamentous, off white and, smooth. (Amin et al., 2014) Whereas, Micrococus were gram positive and non motile. Their colonies on nutrient agar (NA) were circular, convex, Undulate, off white, smooth and spherical (Wesley et al., 1974). Pseudomonas were observed as gram negative and non-motile. Their colonies on nutrient agar (NA) were large, spindle, convex, undulate, yellow, smooth and round. (Mera and Balabasker, 2012). Bascillus were found to be gram positive and non motile. Their colonies on nutrient agar (NA) were large, irregular, raised, lactate, off white, smooth and spherical (Viayalakshmi et al., 2012) (Table 1).
Biochemical characterization
Gram staining, Catalase test and Amylase Tests: Four isolates (67%) namely BRS1, BRS2, BRS3 and BRS4 were found to be Gram positive and two isolates, (33%) namely BRS5 and BRS6 were found to be Gram negative. Out of six, all rhizobacterial isolates (100) were found catalase positive while none was found catalase negative. All rhizobacterial isolates were found to be negative for amylase production also. These results are in accordance with (Joint, 2016) who found all his bacteria positive as well as negative for catalase and amylase production (Table 2).
Table 2: Catalase and amylase response of bacterial isolates.
| Strain No. | Catalase Test (+/-) | Amylase Test (+/-) |
| BRS 15 | + | - |
| BRS 24 | + | - |
| BRS 3 | + | - |
| BRS 4 | + | - |
| BRS 9 | + | - |
| BRS 6 | + | - |
Salt and pH tolerance: Out of six isolates, five rhizobacteria (83%) were found to be positive and one (17%) was found to be negative for 1% salt whereas four rhizobacteria (80%) were found to be positive and two (20%) were found to be negative for 3% salt tolerance. Three rhizobacteria (50%) were found to be positive and three (50%) were found to be negative on 5% salt tolerance. As for as resistivity to pH is concerned, three rhizobacteria (50%) were unable to grow at any pH level. One, among six rhizobacterial strains was found able to grow on 6.5 pH and one on pH 8. Our results are also in accordance with (Naqvi et al., 2016) who successfully grown his culture at 10-450C at 4-9 pH level (Table 3).
Table 3: Salt and Ph tolerance.
| Salt NaCl | pH | NA | |||||
| Strain No | 1% | 3% | 5 % | 6.5 | 7.5 | 8 | Control |
| BRS 15 | + | + | + | + | + | + | + |
| BRS 24 | + | + | + | - | - | + | + |
| BRS 3 | + | + | + | - | - | - | + |
| BRS 4 | + | + | - | - | - | - | + |
| BRS 9 | + | - | - | - | - | - | + |
| BRS 6 | - | - | - | + | - | - | + |
Antagonistic test between rhizobacteria and Fusarium oysporum f. sp. cepae
Antagonistic activity appeared at different magnitudes. All rhizobacterial isolates significantly inhibited the growth of pathogens while the inhibition zone varied from 4.09 to 74.97%. Isolate BR15 and BR4 were found most efficient in in-vitro conditions and exhibited 50.82% and 74.97% inhibition of Fussarium oxysporum f. sp. cepae respectively. The highest (74.97%) inhibitory effect was found in BRS4 while the lowest 4.09 % inhibitory effect was found in BRS3. The plates served as control were found completely covered by fungal mycelia showing no inhibition zone. Mean mycelial inhibition/retardation of the efficient rhizobacterial strain showed that growth inhibition was highly significant at (p < 0.05) as presented in (Table 3). The present study has demonstrated the antagonistic potential of rhizosphere bacterial isolates against Fusarium oysporum f. sp. cepae. that corresponds with previous research works (Dawwam et al., 2013; Muminah et al., 2015) in which they reported the in vitro suppression of plant pathogens by rhizosphere bacterial isolates. In addition, in our study it was shown that some of the rhizobacterial isolates showed little inhibitory activity against Fusarium oysporum f. sp. cepae. (Table 4). This result is in agreement with the previous findings of Ryan et al. (2004). This suggests that the mode of action exerted and/or the type of antibacterial activity produced by the bacterial isolates may vary and that the rhizobacterial isolates are taxonomically different from each other. It has also been reported that fungal diseases can be controlled using antagonistic microbes (Ryan et al., 2004; Hammami et al., 2012).
Table 4: antagonistic activity of rhizobacterial isolates against fungi.
| Strain No. | Fussarium oxysporum | |
| Mycelial growth (mm) | Inhibition over control (%) | |
| BRS 15 |
20.00±0.00BC |
50.82±0.41C |
| BRS 24 |
21.67±0.34B |
45.45±0.45D |
| BRS 3 |
22.00±0.24ABC |
4.09±0.63B |
| BRS 4 |
5.74±0.18D |
74.97±0.71A |
| BRS 9 |
40.34±0.34A |
1.10±0.28F |
| BRS 6 |
21.87±0.18ABC |
4.66±0.74B |
Investigation for further characterization and molecular identification of these soil bacteria and their application in in-vivo is also needed, because if these bacteria are found well for other characteristics then these can be used as biocontrol agents in any disease management strategy. Diseases can be controlled or suppressed by antibacterial or antifungal secretions or through induced systematic resistance. Pseudomonads and Bacillus strains have been reported to be genetically modified and can enhance plant growth and increase the disease resistance in almost all agronomic crops. These rhizobacteria are often dressed on the seed coats before sowing. These inoculated/dressed seeds can adhere enough rhizobacterial populations which makes satisfactory and valuable symbiotic relationship with plant roots and also releases harmful secretions against deleterious microorganisms. Disease management using plant growth promoting rhizobacteria has also been reported as an efficient and attractive tactic in bio-control strategies (Raaijmakers et al., 2002). Many efforts have been made to concentrate on the use of Gram-negative bacteria such as Erwinia or Pseudomonas to manage the crop diseases (Cartwright et al., 1995; Braun-Kiewnick et al., 2000; Shoda, 2000).
Author’s Contribution
Javed Asghar Tariq developed basic ideas, conducted experiments & wrote manuscript. Bashir Ahmed contributed in execution of experiments. Manzoor Ahmed Abro was involved in designing the experiments. M.Ismail & M.Usman helped in writing the manuscript. Raza Muhammad helped in all research activities and revision of manuscript.
References
Amin, M., R. Zeinab., Z.A. Amanollah. 2014. Isolation and identification of Bacillus species from soil and evaluation of their antibacterial properties. Avicenna J. Clin. Microb. Infect. 2(1): e23233.
Braun-Kiewnick, A., B.J. Jacobsen and D.C. Sands. 2000. Biological control of Pseudomonas syringae p.v syringae, the causal agent of basal kernel blight of barley, by antagonistic Pantoea agglomerans. Phytopathol. 90: 368-375. https://doi.org/10.1094/PHYTO.2000.90.4.368
Bakker, P.A.H.M., J.M. Raaijmakers, G.V. Bloemberg, M. Hofte, P. Lemanceau and M. Cooke. 2007. New perspectives and approaches in plant growth-promoting rhizobacteria research. Eur. J. Plant Pathol. 119: 241-242. https://doi.org/10.1007/s10658-007-9114-z
Cartwright, D.K., W.S. Chilton and D.M. Benson. 1995. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotech. 43: 211-216. https://doi.org/10.1007/BF00172814
Dawwam, G.E., A. Elbeltagy, H.M. Emara, I.H. Abbas and M.M. Hassan. 2013. Beneficial effect of plant growth promoting bacteria isolated from the roots of potato plant. Ann. Agric. Sci. 58: 195-201. https://doi.org/10.1016/j.aoas.2013.07.007
De Oliveira, A.N., L. A. de Oliveira., J. S. Andrade and A. F. C. Junior. 2007. Rhizobia amylase production using various starchy substances as carbon substrates. Brazilian J. Microbiol. 38:208-216.
Farzana, Y., R.O.S. Saad and S. Kamaruzaman. 2009. Growth and storage root development of sweet potato inoculated with rhizobacteria under glasshouse conditions. Austral. J. Basic Appl. Sci. 3(Suppl 2): 1461-1466.
Gupta, C.D., R.C. Dubey, S.C. Kang and D.K. Maheshwari. 2001. Antibiotic mediated necrotrophice effect of Pseudomonas GRC2 against two fungal plant pathogens. Curr. Sci. 81: 994.
Hasani, A., A. Kariminik and K. Issazadeh. 2014. Streptomycetes: characteristics and their antimicrobial activities. Int. J. Adv. Bio. Biomed. Res. 2(1): 63-75.
Hammami, I., B. Jaouadi, A.B. Bacha, A. Rebai, S. Bejar, X. Nesme and A. Rhouma. 2012. Bacillus subtilis bacteriocin Bac 14B with a broad inhibitory spectrum: purification, amino acid sequence analysis and physicochemical characterization. Biotechnol. Bioprocess Eng. 17: 41-49. https://doi.org/10.1007/s12257-010-0401-8
James, L.R. 1981. Measurement of personality via conditional reasoning. Org. Res. Methods.1: 131-163. https://doi.org/10.1177/109442819812001
Kennedy, A.C. 2005. Principles and applications of soil microbiology. 2nd Edn. Pearson prentice hall, upper saddle river. N. J. 242-262.
Kloepper, J.W., R. Lifshitz and R.M. Zablotwicz. 1989. Free-living bacterial inocula for enhancing crop productivity. Trend Biotech. 7.
Kloepper, J.W. and M.N. Schroth. 1978. Plant growth-promoting rhizobacteria radishes. In proceedings of the 4th international conference on plant pathogenic bacteria. (2). Station de PathologieVe ´ge ´tale et de Phytobacte ´riologie, INRA, Angers. France. 879-882.
Lazarovits, G. and J. Nowak. 1997. Rhizobacteria for improvement of plant growth and establishment. Hort. Sci. 32: 188-192. https://doi.org/10.21273/HORTSCI.32.2.188
Lynch, J.M. 1990. Introduction: some consequences of microbial rhizosphere competence for plant and soil. In: Lynch J.M. (ed) The rhizosphere. Wiley, Chichester. 1-10.
Meera, T. and P. Balabaskar. 2012. Isolation and characterization of pseudomonas fluorescens from rice fields. Int. J. Food Agric. Vet. Sci. 2(1): 113- 120.
Merina, P.D., P.D. Vennila and Y. Yasmine. 2015. A study on antagonistic potential of bacteria against phytopathogenic. Fungi. Int. J. Pharm. 34(1): 191-193.
Muminah, B., H. Subair and Fahruddin. 2015. Isolation and screening of bacterial exopolysaccharide (EPS) from potato rhizosphere in highland and their potential as a producer of Indole Acetic Acid (IAA). Procedia Food Sci. 3: 74-81. https://doi.org/10.1016/j.profoo.2015.01.007
Munis and N. Mulugeta. 2014. Field management of anthracnose (Colletotrichum lindemuthianum) in common bean through fungicides and bioagents. Adv. Crop Sci. Tech. 2: 124.
Naqvi, S. A. H., R. Perveen., U. D. Umar. A. U. Rehman., S. Chohan and S. H. Abbas. 2016. Bacterial leaf blight of rice a disease forecasting model based on meteorological factors in Multan. Pakistan. J. Agric. Res. 54 (4): 707-718.
Jonit, N.Q., Y.C. Low and G.H. Tan. 2016. Xanthomonas oryzae pv. oryzae, Biochemical tests, rice (Oryza sativa). Bacterial leaf blight (BLB) Disease. Sekinchan J. Appl. Environ. Microbiol. 4(3): 63-69.
Raaijmakers, Vlami, J.M. Souza and M. JT. 2002. Antibiotic production by bacterial biocontrol agents. Anton van Leeuwenhoek. 81: 537-547. https://doi.org/10.1023/A:1020501420831
Ryan, A.D., L.L. Kinkel and J.L. Schottel. 2004. Effect of pathogen isolate, potato cultivar and antagonist strain on potato scab severity and biological control. Biocont. Sci. Technol. 14: 301-311. https://doi.org/10.1080/09583150410001665187
Suresh, S.R., S.T. Yenjerappa, M.K. Naik, S.B. Mallesh and C.M. Kalibavi. 2013. Studies on cultural and physiological characters of Xanthomonas oryzae pv. oryzae causing bacterial blight of rice. Karnataka J. Agric. Sci. 26(2): 214-216.
Serge, S., F. Andra and N.A. Jaen. 2012. Crop losses due to diseases and their implications for global food production losses and food security. Food Sec. DOI.10-1007/S 12571. Springer Science + Bus. Media Int. Soc. Plant Pathol.
Shoda. 2000. Bacterial control of plant diseases. J. Biosci. Bioeng. 89(6): 515-521.
Tilak, K.V.B.R., N. Ranganayak, K.K. Pal, A. Kxena, C.S. Nautiyal, S. Mittal, A.K. Tripathi and B.N. Johri. 2005. Diversity of plant growth and soil health supporting bacteria. Curr. Scie. 89 (Suppl 1): 136-150.
Van Loon and Bakker. 2003. Plant responses to plant growth-promoting rhizobacteria. Eur. Pl. Pathol. 119: 243-254.
Van Loon and Glick. 2004. Increase plant fitness by rhizobacteria in; Sandermann. H, ed. Molecular ecotoxicology of plant. Ecol. study. 170: 177-205. https://doi.org/10.1007/978-3-662-08818-0_7
Vijayalakshmi, K. Sushma, S. Abha and P.I. Chander. 2012. Isolation and characterization of Bacillus subtilis KC3 for amylolytic activity. Int. J. Biosci. Biochem. Bioinform. 2(5): 2012.
Vincent, J.M. 1947. Distortion of fungal hyphae in presence of certain inhibitors. Nat. 159: 850. https://doi.org/10.1038/159850b0
Wesley, K.E., T. Tornabene and H.S. Karl. 1974. Isolation and characterization of micro cocci from human skin, including two new species: Micrococcus zyzae and Micrococcus kristinae. Int. J. Syst. Bacteriol. 79-109. Int. Assoc. Microbio. Societ. 24(1).
Zaman, A., F.A. Hossen, M. Razi, M. Ismail A. Hoque, M. Zahurul-Islam, S.M. Shahidullah and S. Meon. 2009. Efficiency of plant growth-promoting rhizobacteria (PGPR) for the enhancement of rice growth. Afr. J. Biotech. 8 (7): 1247-1252.
To share on other social networks, click on any share button. What are these?







