Molecular Identification of Bovine Alpha Herpesvirus Subtype 1.1 in Field Isolates in Egypt by Phylogenetic Analysis of Glycoprotein B Gene
Molecular Identification of Bovine Alpha Herpesvirus Subtype 1.1 in Field Isolates in Egypt by Phylogenetic Analysis of Glycoprotein B Gene
Basel A. Abokhadra, Samah M. Mosad, Sahar Abd El Rahman*
Clinically diseased animals suspected to be infected with BoHV-1 showing: (A) Red nose and nasal discharge; (B) open mouth breathing; (C) Corneal opacity.
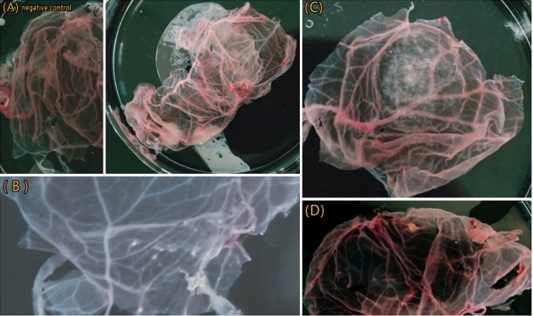
Results of BoHV-1 isolation on CAM showing. (A) Control negative and CAM of 1st passage showing thickening and congestion. (B) CAM of 2nd passage showing Small number of pocks. (C) CAM of 3rd passage showed typical pock lesion. (D) CAM of 3rd passage showed small and large pock lesion.
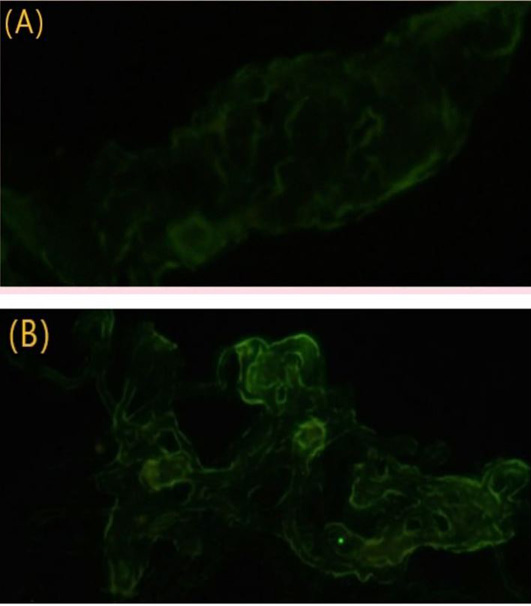
Result of IFT from CAMs. (A) Control negative; (B) CAM from the third passage shows yellowish-green fluorescence.
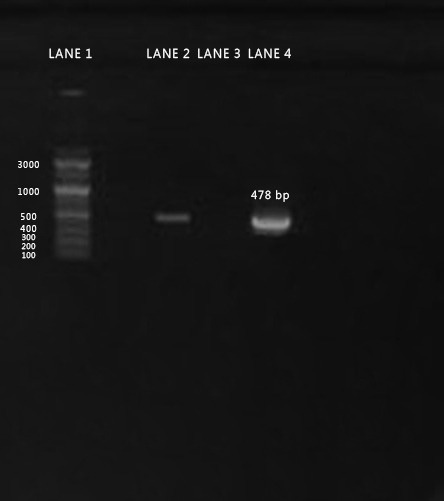
Agarose gel electrophoresis of PCR amplicon for gB gene of BoHV-1isolate separated on 1% agarose gel and stained with ethidium bromide (Positive PCR amplicons are at the size of approximately 478 bp). Lane 1 denotes for 100 bp DNA Ladder, Lane 2: Positive sample, Lane 3 negative control. Lane 4: positive control.
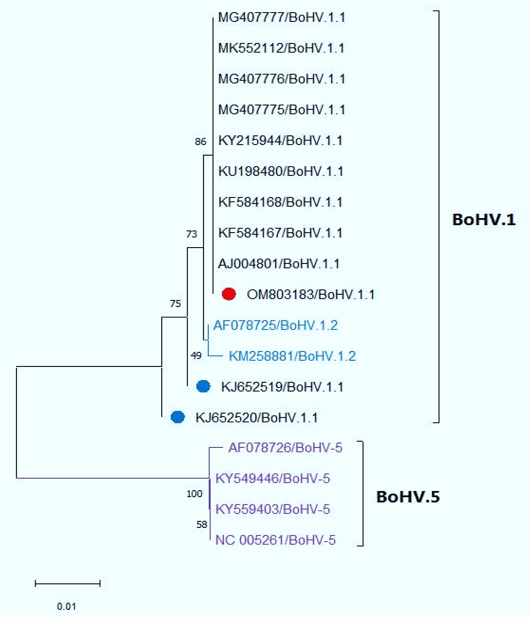
Neighbor-Joining tree was constructed based on partial sequencing of the gB gene showing the genetic relatedness of our Egyptian isolate (OM803183) to other reference strains from GenBank. Our isolate showed 100% homology to BoHV1.1 reference strains Including Egyptian BoHV-1.1 reference strains (blue circle) with query coverage 63% but away from BoHV-5, which are located in a separate clade.
Nucleotide sequence alignment of our isolate in comparison to other strains from Genbank with AJ004801 as reference strain using Bioedit software version 7.1
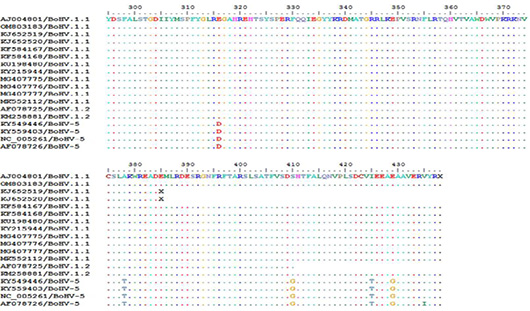
Deduced amino acid sequence alignment of our isolate in comparison to other strains from Genbank with AJ004801 as reference strain using Bioedit software version 7.1