Effect of Colebrookea oppositifolia Leaf Extracts on Cytochrome P450 and Glutathione S-Transferase Activity in Rats
Research Article
Effect of Colebrookea oppositifolia Leaf Extracts on Cytochrome P450 and Glutathione S-Transferase Activity in Rats
Rukhsana Anwar1*, Ammara Abd-Us-Sattar1, Afifa Noor2, Kanwal Ashiq1,3 and Shah Jahan4
1Punjab University College of Pharmacy, University of the Punjab Lahore, Pakistan; 2Institute of Microbiology and Molecular Genetics, University of the Punjab Lahore, Pakistan; 3Faculty of Pharmaceutical Sciences, Superior University Lahore, Pakistan; 4Department of Immunology, University of Health Sciences, Lahore Pakistan.
Abstract | Colebrookea oppositifolia is a medicinal plant that contains many therapeutic constituents. Till now, research has not been conducted related to this plant’s effect on hepatic drug-metabolizing enzymes and glutathion S-transferase. For this purpose, different cytochrome (CYP450) modulators were selected to determine pharmacokinetic interactions. Cimetidine, rifampicin, dexamethasone, and tamoxifen were selected as CYP450 modulators. In a sub-acute in vivo study, 200 mg/kg BW of both extracts were orally administered once daily for 19 days to different groups of rats. CYP modulators were administered to respective groups for last 5 consecutive days. Real-time PCR analysis of hepatic CYP450 subtypes, CYP1A2, CYP2C9, and CYP3A4 was carried out. Moreover, phase II drug-metabolizing enzymes; GST-specific activity, total glutathione, superoxide dismutase, catalase, and malondialdehyde were determined in liver homogenates. The phytochemical analysis of extracts revealed the presence of many important bioactive compounds. Real-time PCR results showed that aqueous extract decreased the mRNA expression of CYP1A2, CYP3A4. The methanolic extract decreased mRNA expression of CYP2C9 and CYP3A4. Both extracts enhanced GST-specific activity, tGSH, SOD, and CAT activity in rats. It is concluded that leaf extracts of C. oppositifolia showed significant interaction with drugs. This can modulate drug-metabolizing enzyme activity and reduce oxidative stress.
Received | October 15, 2021; Accepted | June 10, 2022; Published | June 20, 2022
*Correspondence | Rukhsana Anwar, Punjab University College of Pharmacy, University of the Punjab Lahore, Pakistan; Email: rukhsana.pharmacy@pu.edu.pk
Citation | Anwar, R., A.A. Sattar, A. Noor, K. Ashiq and S. Jahan. 2022. Effect of Colebrookea oppositifolia leaf extracts on cytochrome P450 and glutathione S-transferase activity in rats. Biologia (Lahore), 68(1): 1-10.
DOI | https://dx.doi.org/10.17582/journal.Biologia/2022/68.1.1.10
Keywords | Colebrookea oppositifolia, mRNA, CYP, Pharmacokinetic interaction, Enzyme, Medicinal plant
Copyright: 2022 by the authors. Licensee ResearchersLinks Ltd, England, UK.
This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Introduction
Colebrookea oppositifolia is a genus of the Lamiaceae family. Its common name is Bhaman. It has therapeutic attributes due to the presence of polyphenols and flavonoids in abundance. It has anti-inflammatory, antimicrobial, anthelmintic, hepatoprotective, antioxidant, anti-hypertensive, and antidiabetic activities. Due to its anti-inflammatory activity, it is used in the treatment of conjunctivitis (Shirsat et al., 2014; Viswanatha et al., 2021). Leaves and roots of C. oppositifolia have wound healing and antiepileptic activity, respectively (Ajaib et al., 2011). The plant leaves contain 4, 5, 6, 7-tetra methoxy flavones, 5, 6, 7 trimethoxyflavones, Catechin -7-O-β rhamnopyranoside, and quercetin (Madhavan et al., 2011; Yadav, 2019). According to a study, acteoside was isolated in a pure form from the C. oppositifolia and this compound exhibited hepatoprotective activity (Sardar and Manik, 2017).
When the effect of one drug is altered by any other xenobiotic that may be a drug product, drink, food, or any chemical agent, this phenomenon is known as drug interaction (Baxter and Preston, 2010; Babos et al., 2021). The pharmacokinetic interactions can be modified by either increasing or decreasing the concentration of the drug at the receptor site. These interactions may be due to the inhibition or induction of the enzymes that are involved in drug metabolism (Li et al., 2021). Further, drug competition for the uptake by transporter can also influence the rate of drug metabolism. For example, ciprofloxacin is an inhibitor of CYP1A2 and it also metabolizes theophylline. Therefore, when ciprofloxacin is used in combination with theophylline, the plasma concentration of theophylline will increase and could cause gastrointestinal and cardiac side effects (Cascorb, 2012; Chappell et al., 2020). Pharmacodynamic interactions occur when another drug is present at the target site. These may be either antagonistic interactions or synergistic interactions. For example, when ethanol is used in combination with the benzodiazepine it is synergistic interaction and when flumazenil is given in combination with the benzodiazepine it is an antagonistic interaction (Anwar et al., 2012).
Enzymes of drug metabolism play an important role in biotransformation. Moreover, they are involved in the detoxification of xenobiotics and thus provide a defense to the body against harmful chemicals. Enzymes of drug metabolism are divided into two phases, including phase I enzymes and phase II enzymes. Microsomal enzymes are present in the lungs, liver, gastrointestinal tract, and kidneys. Phase II enzymes are also known as conjugating enzymes and these include menadione reductase, sulfotransferases, glutathione S-transferases, epoxide hydrolases, and N-acetyl transferases. These enzymes enhance the hydrophilicity of xenobiotics and thus increase the excretion of xenobiotics (Rushmore and Tony, 2002; Kim et al., 2021).
Many drugs can modulate the effect of different cytochromes that involve in the regulation of the drug metabolism. For instance, cimetidine is an inhibitor of CYP1A2, CYP2C9, and CYP3A4, rifampicin acts as an inducer of CYP3A4, CYP1A2, CYP2C9, and CYP2D6, dexamethasone is the inducer of CYP3A4 and CYP2D6, and tamoxifen is the substrate of CYP2D6 and CYP3A4 (Martínez et al., 1999; Lin et al., 1999; Crewe et al., 2002). The disturbance in the balance between the reactive oxygen species such as peroxide, superoxide, hydroxyl, hydrogen peroxide, peroxyl radicals, and the antioxidant defense system may lead to oxidative stress (Mushtaq et al., 2018). Malondialdehyde, catalase, and total glutathione are markers of oxidative stress. MDA levels will increase while CAT and GSH quantities will decrease during oxidative stress (Mushtaq et al., 2020).
So far, the plant’s effect on hepatic drug-metabolizing enzymes cytochrome P450 and glutathiones-transfers has not been investigated. For this purpose, different CYP450 modulators (cimetidine, rifampicin, dexamethasone, and tamoxifen) were selected to assess pharmacokinetic interactions.
Materials and Methods
Chemicals
1-Cloror-2, 4-dinitrobenzene (CDNB), glutathione (GSH), potassium dichromate, potassium sodium tartrate, pyrogallol, sodium hydroxide, 1, 1, 3, 3-Tetramethoxy propane were supplied by Sigma-Aldrich. Copper sulfate, dipotassium hydrogen phosphate, glacial acetic acid, hydrochloric acid and methanol were supplied by Merck. G script first-strand synthesis kit, total RNA isolation kit and Eva Green qPCR master mix were purchased from gene Direx.
Animals
Male sprague-dawley rats that had a weight range of 150-250g were used for in vivo study and purchased from the University of Veterinary and Animal Sciences, Lahore, Pakistan. Guidelines of the PUCP animal ethics committee were followed for the handling of animals. The experimental protocol of the current study was approved by the PUCP Animal Ethics Committee and the voucher number was assigned (AEC/PUCP/1096). Rats were housed in plastic cages with top of steel mesh in the animal house of Punjab University College of Pharmacy under adequate environmental conditions. They were kept under observation before the experiment and provided with adequate diet and water.
Preparation of extracts
Fresh leaves of C. oppositifolia were collected from Haripur (a district of Khyber Pakhtunkhwa). The identification of plants was done by Dr. Zaheer (Professor, Department of the Botany, GC University, Lahore) and also, he issued the voucher number GC.Herb.BOT.2973. Specimen of the voucher deposited at the Herbarium Department of GC University Lahore, Pakistan. The leaves of C. oppositifolia were first separated from the stem, washed, and air-dried. Then, the dried leaves were grounded to powder. The maceration method was used for the preparation of aqueous and methanolic extract of C. oppositifolia, and the percentage yield was calculated to be 4.4% and 8.25%, respectively.
Experimental design
Ninety male Sprague Dawley rats were divided into fifteen groups. In each group, six rats (n=6) were included. The detail of each group is given following.
Group I (control)
The vehicle only for 19 days.
Group II and III
Aqueous and methanolic extract (200 mg/kg) administered orally for 19 days.
Group IV
Cimetidine (20mg/kg) administered orally for the last five consecutive days.
Group V and VI (Aqueous and methanolic extract + Cimetidine)
The 200 mg/kg of extract was given orally for 14 days, and cimetidine was co-administered for the last five consecutive days.
Group VII
Rifampicin (50 mg/kg) administered orally for the last five consecutive days.
Group VIII and IX (Aqueous and methanolic extract + Rifampicin)
The 200 mg/kg of extract was administered orally for 14 days, and rifampicin was co-administered for the last 5 consecutive days.
Group X
Dexamethasone (40 mg/kg) orally for the last 5 consecutive days.
Group XI and XII (Aqueous and methanolic extract + Dexamethasone)
The 200 mg/kg of extract was administered orally for 14 days, and dexamethasone was co-administered for the last five consecutive days.
Group XIII
Tamoxifen (5mg/kg) had given orally for the last five consecutive days.
Group XIV and XV (Aqueous and methanolic extract + Tamoxifen)
The 200 mg/kg of extract orally for 14 days, and tamoxifen was co-administered for the last five consecutive days.
The rats were sacrificed on the 20th day and their livers were separated and preserved in the chiller at -80°C.
Standardization of extracts
The standardization of aqueous and methanolic extract of C. oppositifolia was done to determine the concentration of total flavonoids, total polyphenols, total polysaccharides, total protein and glycosaponins in both extracts.
Total flavonoids
The method of Chang was utilized for the quantification of flavonoids in the extracts with a slight alteration. The Standard used was quercetin. A stock solution (1mg/1ml) of quercetin and extracts was prepared in methanol. Different dilutions of quercetin (10, 20, 40, 80, 100, and 120 µg/ml) were made. The final reaction mixture containing 200 µl of the sample/standard, 10% w/v of aluminum nitrate solution, and 1M potassium acetate was incubated for 45 minutes. Absorbance was measured at 415 nm. The calibration curve was plotted for the determination of total flavonoids (expressed as mg/g of quercetin) (Chang et al., 2002).
Total polyphenols
The method of Slinkard and Singleton was utilized for the quantification of total polyphenols in both extracts with a slight alteration. The Standard used was gallic acid. Stock solution (1mg/1ml) of gallic acid and extracts was prepared in methanol. Different dilutions of gallic acid (10, 20, 40, 80, 100, and 120 µg/ml) were made. The final reaction mixture containing 200 µl of the sample/standard, FC reagent, sodium carbonate was incubated at room temperature for 2 hours then absorbance was measured at 760 nm. The calibration curve was plotted for the determination of total polyphenol (expressed as mg/g of gallic acid) (Slinkard and Singleton, 1977).
Total protein
The method of Lowry was utilized for the quantification of total protein content in extracts with a slight alteration. Both extracts (50 mg) were dissolved in 10 ml of the distilled water then 5 drops of the Triton-X were added. After centrifugation supernatant was separated and volume was made up to 1ml by distilled water. Then 3ml of the reagent C was added. Reagent C was prepared by the mixing of 50 ml of reagent A (2% of the Na2CO3 in 0.1 N solution of NaOH) and one ml of reagent B (0.5% of the Copper sulfate in 1% of potassium sodium tartrate). Then 200 µl of the FC reagent was added and incubated at room temperature for 30 minutes. Absorbance was measured at 600 nm. The Standard used as bovine serum albumin and its 1mg/1ml stock solution was prepared then dilutions (10, 20, 40, 60, 80, and 100 µg/ml) were made. The calibration curve was plotted for the determination of total protein content in both extracts (Lowry et al., 1951).
Total glycosaponins
The method of Hussain was utilized for the quantification of glycosaponins in both extracts with a slight alteration. Extracts were refluxed with methanol for 30 minutes. Filtration was done the filtrate obtained was concentrated on a water bath then added in acetone. The precipitates formed were dried at 100 oC and weighed then the following formula was used to determine the content of glycosaponins in both of the extracts (Hussain et al., 2008).
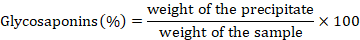
Carbohydrates
Firstly 50 mg of each extract dissolved in hot ethanol and vortexed for 2 minutes. After centrifugation supernatant was collected. Further, extraction of supernatant was carried out with hydrochloric acid and distilled water in a ratio of 1:1. The supernatant was collected and extraction repeated two times. The reaction mixture containing 100 µl of supernatant, 900 µl water, and 4 ml of the anthrone reagent was placed in a boiling water bath for 8 minutes then cooled. The green color was produced. Its absorbance was measured at 630 nm. Glucose was used as standard and a calibration curve was plotted (Yemm and Willis, 1954).
RT-PCR analysis
Gene direx total RNA isolation kit was used for the extraction of RNA according to the manufacturer guidelines. The purity of RNA was determined by Nano-drop and then RNA pellet was preserved in the chiller at -80°C for further use. G script first-strand synthesis kit by gene direx was used for the synthesis of cDNA and cDNA was synthesized according to the manufacturer guidelines. The resulting reverse transcription product was preserved in the chiller at -80°C for further use.
Eva Green qPCR master mix was thawed and then the reaction was performed with a final volume of 20 µl and this volume contained 10 µl of Eva Green qPCR master mix 1 µl of cDNA, 0.5 µl of forward and reverse primer, and 9 µl of Nuclease free water. The reaction was performed in 96 well RT-PCR plates. Real-time PCR analysis was carried out with initial denaturation for 5 minutes at 95 °C and then annealing for 60 seconds at 60°C followed by extension for 60 seconds at 60°C. The sequence of forward and reverse primers used were as follows:
|
Isoenzymes |
Primers |
|
CYP1A2 |
F:5’-TCAACCTCGTGAAGAGCAGCA-3’ R: 5’-CCGAAGAGCATCACCTTCTC-3’ |
|
CYP2C9 |
F:5’-AAAAGCACAATCCGCAGTCT-3’ R:5’-GCATCTGGCTCCTGTCTTTC-3’ |
|
CYP3A4 |
F:5’-TCTGTGCAGAAGCATCGAGTG-3’ R:5’-TGGGAGGTGCCTTATTGGG-3’ |
|
CYP2E1 |
F:5’-CCTACATGGATGCTGTGGTG-3’ R:5’-CTGGAAACTCATGGCTGTCA-3’ |
F: Forward primer; R: Reverse primer.
Glutathione S-transferase assay
The method of Habig et al. (1974) was utilized for the estimation of GST activity in liver tissue with a slight alteration. The assay was performed in three replicates. Phosphate buffer, GSH, and CDNB were added in the test tubes containing post mitochondrial supernatant respectively. Blank contained all the components except supernatant. Change in the absorbance for one minute interval was measured at 340 nm on spectrophotometer up to 3 minutes. GST activity was expressed in μmol/ minute/ mg protein (Anwar et al., 2012). The following formula was used for estimating GST activity.

Total glutathione assay
Total glutathione level in liver tissue of rats was quantified by the method of Sedlak and Lindsay with a slight alteration. Liver tissue was weighed and phosphate buffer was added to it. Liver tissue was homogenized with phosphate buffer then 25% trichloroacetic acid was added to precipitate the homogenate. It was centrifuged for forty minutes. The clear supernatant was separated. The final reaction mixture contained, Tris-HCl buffer, supernatant, DTNB, and methanol. Blank was prepared by the same method except that phosphate buffer was used in place of supernatant. All contents vortexed then incubated for thirty minutes at 37°C. The yellow color appeared and its absorbance was measured at 412 nm (Sedlak and Lindsay, 1968).
Catalase assay
The Sinha method was used for the determination of catalase activity in liver tissue with a slight alteration. The test was performed by making three replicates of each sample. 0.01M phosphate buffer, 0.02M hydrogen peroxide were added homogenate. All contents were vortexed. One ml of this mixture was taken then a reagent was added which resulted in blue-colored precipitates. The reagent was made by mixing potassium dichromate and glacial acetic acid in a ratio of 1:3. All contents were vortexed then heated at 100°C for ten minutes. Blue-colored precipitates changed to green-colored solution. Absorbance was measured at 570 nm (Sinha, 1972; Anwar et al., 2012).
Malondialdehyde assay (MDA)
The Ohkawa method was utilized for the determination of MDA level in liver tissue with a little alteration. Liver tissues were homogenized in 1.15% potassium chloride. The test was performed by making three replicates of each sample. The reaction mixture containing 8% sodium lauryl sulfate, 20% acetic acid, 0.8% thiobarbituric acid, distilled water, and 10% homogenate was incubated at 98°C for one hour then cooled and at the last n-butanol was added. The whole contents were vortexed then centrifuged for thirty minutes at 4000 rpm. Blank was prepared by using potassium chloride in place of homogenate. Absorbance was measured at 532 nm (Ohkawa et al., 1979; Anwar et al., 2012).
Superoxide dismutase (SOD) assay
The Magnani method was utilized for the estimation of superoxide dismutase levels in liver tissue of rats with a slight alteration. The liver homogenate was prepared in 67mM phosphate buffer (pH7.4). The reaction mixture contained 1 ml of Tris-buffer, homogenate, and 0.2mM pyrogallol solution was added just before the determination of the absorbance. In control distilled water was used in place of the liver homogenate. The Tris-buffer was used as blank. Absorbance was measured at 420 nm. The following formula was used for the calculation of superoxide dismutase activity (Magnani et al., 2000; Ijaz et al., 2021).

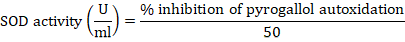
Statistical analysis
Graph pad Prism version 5 was used for statistical analysis. One-way ANOVA and Dunnett’s multiple comparison test was used for making a comparison between different groups. Results were expressed in mean and standard deviation (Mean±SD) and *p < 0.05 was considered statistically significant.
Results and Discussion
Standardization quantified in aqueous and methanolic extract, respectively were total flavonoids (85.95 mg/g and 93.86 mg/g), polyphenols (47.69 mg/g and 39.23 mg/g), carbohydrates (69.96 mg/g and 66.14 mg/g), protein (80.32 mg/g and 89.57 mg/g) and glycosaponins (17.6% and 10.4%), respectively as shown in Table 1.
RT-PCR analysis was carried out for the determination of mRNA expression of CYP1A2, CYP2C9, and CYP3A4 and results showed that aqueous extract significantly decreased (*p < 0.05) the mRNA expression of CYP1A2, and CYP3A4 by 0.88-fold, and 0.78-fold, respectively however, it showed no significant effect on mRNA expression of CYP2C9 as compared to the control. The methanolic
Table 1: Standardization results of aqueous and methanolic extract of Colebrookea oppositifolia.
|
Extracts |
Total flavonoids content (mg/g) |
Total polyphenols content (mg/g) |
Carbohydrate content (mg/g) |
Total protein content (mg/g) |
Total glycosaponins content (%) |
|
Aqueous extract |
85.95 ± 1.94 |
47.692 ± 2.308 |
69.96 ± 3.96 |
80.32 ± 2.23 |
17.6 ± 1.6 |
|
Methanolic extract |
92.95 ± 3.71 |
39.230 ± 1.538 |
66.14 ± 7.22 |
89.57 ± 3.36 |
10.4 ± 2.4 |
extract significantly decreased (*p < 0.05) the mRNA expression of CYP2C9 and CYP3A4 by 0.87-fold and 0.69-fold, respectively but it showed no significant effect on mRNA expression of CYP1A2 as compared to the control as shown in Table 2.
Table 2: effect of aqueous and methanolic extracts on Expression of CYPs.
|
Groups |
CYP1A2 |
CYP2C9 |
CYP3A4 |
|
Control |
1.03 ± 0.078 |
1.03 ± 0.078 |
1.03 ± 0.078 |
|
Aqueous extract |
0.88 ± 0.070* |
0.98 ± 0.041 |
0.78 ± 0.049* |
|
Methanolic extract |
0.90 ± 0.028 |
0.87 ± 0.062* |
0.69 ± 0.036* |
Cimetidine significantly decreased (*ap < 0.05) the mRNA expression CYP1A2, CYP2C9 and CYP3A4 by 0.76-fold, 0.66-fold, and 0.56-fold, respectively as compared to the control. Aqueous extract co-administered with cimetidine significantly decreased (*p < 0.05) the mRNA expression of CYP2C9 and CYP3A4 by 0.52-fold and 0.42-fold, respectively; however, it showed no significant effect on CYP1A2 as compared to the cimetidine treated group. The methanolic extract co-administered with cimetidine significantly decreased (*p < 0.05) the mRNA expression of CYP2C9 and CYP3A4 by 0.47-fold and 0.37-fold, respectively.
However, it showed no significant effect on CYP1A2 as compared to the cimetidine treated group as shown in Table 3.
Table 3: Effect of co administration of extracts and cimetidine on CYPS expression.
|
Groups |
CYP1A2 |
CYP2C9 |
CYP3A4 |
|
Control |
1.03±0.078 |
1.03±0.078 |
1.03±0.078 |
|
Cimetidine |
0.76±0.037*a |
0.66±0.041*a |
0.56±0.055*a |
|
Aqueous extract + Cimetidine |
0.82±0.037 |
0.52±0.064* |
0.42±0.045* |
|
Methanolic extract + Cimetidine |
0.77±0.025 |
0.47±0.030* |
0.37±0.063* |
Rifampicin significantly increased (*p < 0.05) the mRNA expression of CYP1A2, CYP2C9 and CYP3A4 by 1.76-fold, 1.66-fold and 1.79-fold, respectively as compared to the control. Aqueous extract co-administered with rifampicin significantly decreased (*ap < 0.05) the mRNA expression of CYP1A2, CYP2C9 and CYP3A4 by 1.66-fold, 1.47-fold and 1.58-fold, respectively as compared to the rifampicin treated group. Methanolic extract co-administered with rifampicin significantly decreased (*p < 0.05) the mRNA expression of CYP1A2 and CYP3A4 by 1.59-fold and 1.45-fold, respectively however, it showed no significant effect on mRNA expression of CYP2C9 as compared to the rifampicin treated group as shown in Table 4.
Table 4: Effect of co administration of extracts and rifampicin on CYPS expression.
|
Groups |
CYP1A2 |
CYP2C9 |
CYP3A4 |
|
Control |
1.03±0.078 |
1.03±0.078 |
1.03±0.078 |
|
Rifampicin |
1.76±0.026*a |
1.66±0.061*a |
1.79±0.045*a |
|
Aqueous extract+ Rifampicin |
1.66±0.040* |
1.47±0.070* |
1.58±0.036* |
|
Methanolic extract + Rifampicin |
1.59±0.026* |
1.54±0.041 |
1.45±0.058* |
Dexamethasone significantly increased (*p < 0.05) the mRNA expression of CYP3A4 by 1.64-fold respectively compared to the control. Aqueous extract co-administered with dexamethasone significantly decreased (*p < 0.05) the mRNA expression of CYP3A4 by 1.37-fold compared to the dexamethasone-treated group. Methanolic extract showed no significant effect on mRNA expression of CYP3A4 compared to the dexamethasone-treated group as shown in Table 5. Tamoxifen showed no significant effect on mRNA expression of CYP3A4 as compared to the control.
Table 5: Effect of coadministration of extracts and dexamethasone on CYP3A4 expression.
|
Groups |
CYP3A4 |
|
Control |
1.03 ± 0.078 |
|
Dexamethasone |
1.64 ± 0.040*a |
|
Aqueous extract + Dexamethasone |
1.37 ± 0.035* |
|
Methanolic extract + Dexamethasone |
1.67 ± 0.056 |
Aqueous extract co-administered with tamoxifen significantly decreased (*p < 0.05) the mRNA expression of CYP3A4 by 0.78-fold as compared to the tamoxifen treated group. Methanolic extract co-administered with tamoxifen showed no significant effect on mRNA expression of CYP3A4 as compared to the tamoxifen treated group as shown in Table 6.
Table 6: Effect of co administration of extracts tamoxifen on CYP3A4 expression.
|
Groups |
CYP3A4 |
|
Control |
1.03 ± 0.078 |
|
Tamoxifen |
1.12 ± 0.065 |
|
Aqueous extract + Tamoxifen |
0.78 ± 0.081* |
|
Methanolic extract + Tamoxifen |
1.17 ± 0.045 |
The results of the following activities; glutathiones transferase (GST), total glutathione (tGSH), catalase (CAT), malonaldehyde (MDA), and superoxide dismutase (SOD) are displayed in Table 7. Both extracts enhanced the GST-specific activity, tGSH, SOD, and CAT activity while decreased the MDA activity in rats. Hence, these extracts could be beneficial in reducing oxidative stress.
Worldwide, medicinal plants are widely used to treat various disorders. Medicinal plants are considered safe, cheap and effective by many people around the globe (Tanveer et al., 2019; Qayyum et al., 2020; Latif et al., 2020; Ashiq et al., 2021).
The current work was conducted on Colebrookea oppositifolia to explore its potential effect on cytochromes P450 and its interaction with other drugs. Standardization results showed that total flavonoids content in aqueous and methanolic extract of C. oppositifolia were 85.95 mg/g and 93.86 mg/g, respectively, total polyphenols were 47.69 mg/g and 39.23 mg/g, respectively, carbohydrates were 69.96 mg/g and 66.14 mg/g, respectively.
One way ANOVA and dunnett’s multiple comparison test was used for conducting data analysis between different groups. Results were expressed in mean and standard deviation (Mean±SD) and *p < 0.05 was considered statistically significant.
Total protein was 80.32 mg/g and 89.57 mg/g respectively and total glycosaponins were 17.6 % and 10.4 % respectively. The plant is an important pool of various chief phytochemicals which confirms the therapeutic significance of the plant (Sharma et al., 2004).
The methanolic extract significantly decreased (*p < 0.05) the mRNA expression of CYP2C9 and CYP3A4 while it showed no significant effect on mRNA expression of CYP1A2 and CYP2E1 compared to the control. Cimetidine inhibited the activity of CYP1A2, CYP2C9 and CYP3A4 by decreasing their mRNA expression and extracts of C. oppositifolia co-administered with cimetidine further decreased their mRNA expression thus exhibited synergistic effect.
Table 7: Herb-drug interaction effect of C. oppositifolia on liver enzymes activity and levels with different drugs.
|
Groups |
GST activity (µM/min/mg protein) |
tGSH (mM/g tissue) |
CAT (µmole of H2O2 consumed/min/mg) |
MDA (µM/g tissue) |
SOD (U/mg) |
|
Control |
0.092 ± 0.001 |
4.202 ± 0.095 |
43.10 ± 1.129 |
1.698 ± 0.101 |
4.689 ± 0.092 |
|
AE |
0.093 ± 0.003 |
4.297 ± 0.103 |
43.13 ± 1.090 |
1.717 ± 0.137 |
4.662 ± 0.178 |
|
ME |
0.089 ± 0.003 |
4.169 ± 0.159 |
43.04 ± 1.196 |
1.714 ± 0.147 |
4.648 ± 0.344 |
|
Cimetidine |
0.063 ± 0.002*a |
1.731 ± 0.146*a |
24.06 ± 1.131*a |
5.661 ± 0.207*a |
1.243 ± 0.139*a |
|
AE+ Cimetidine |
0.082 ± 0.001* |
2.998 ± 0.141* |
34.65 ± 1.667* |
3.220 ± 0.116* |
3.301 ± 0.088* |
|
ME+ Cimetidine |
0.075 ± 0.001* |
2.743 ± 0.179* |
34.14 ± 1.096* |
3.336 ± 0.120* |
3.203 ± 0.115* |
|
Rifampicin |
0.061 ± 0.001*a |
1.617 ± 0.102*a |
23.08 ± 0.990*a |
6.594 ± 0.098*a |
1.486 ± 0.112*a |
|
AE+ Rifampicin |
0.076 ± 0.002* |
2.730 ± 0.123* |
34.03 ± 1.059* |
3.389 ± 0.113* |
2.810 ± 0.132* |
|
ME+ Rifampicin |
0.074 ± 0.001* |
2.639 ± 0.166* |
32.86 ± 0.952* |
3.717 ± 0.108* |
2.396 ± 0.142* |
|
Dexamethasone |
0.062 ± 0.001*a |
1.494 ± 0.111*a |
23.30 ± 2.055*a |
6.265 ± 0.125*a |
1.31 ± 0.092*a |
|
AE+Dexamethasone |
0.081 ± 0.001* |
2.918 ± 0.127* |
33.18 ± 1.482* |
3.713 ± 0.127* |
2.78 ± 0.150* |
|
ME+Dexamethasone |
0.076 ± 0.002* |
2.686 ± 0.124* |
33.00 ± 1.431* |
4.109 ± 0.145* |
2.40 ± 0.174* |
|
Tamoxifen |
0.067 ± 0.002*a |
2.200 ± 0.096*a |
25.66 ± 1.759*a |
5.398 ± 0.123*a |
1.806 ± 0.118*a |
|
AE+ Tamoifen |
0.083 ± 0.002* |
3.299 ± 0.091* |
35.55 ± 1.698* |
3.108 ± 0.110* |
3.500 ± 0.135* |
|
ME+ Tamoxifen |
0.081 ± 0.001* |
3.074 ± 0.134* |
34.80 ± 1.172* |
3.218 ± 0.112* |
3.315 ± 0.186* |
Rifampicin enhanced the activity of CYP1A2, CYP2C9 and CYP3A4 by increasing their mRNA expression and extracts of C. oppositifolia co-administered with rifampicin opposed the effects of rifampicin and decreased the mRNA expression of CYP1A2, CYP2C9 and CYP3A4. Dexamethasone enhanced the activity of CYP3A4 by increasing its mRNA expression and extracts of C. oppositifolia co-administered with dexamethasone opposed its effects and decreased the mRNA expression of CYP3A4. Tamoxifen showed no effect on mRNA expression of CYP3A4; however, extracts of C. oppositifolia co-administered with tamoxifen decreased the mRNA expression of CYP3A4. Aqueous and methanolic extracts co-administered with cimetidine, rifampicin, dexamethasone, and tamoxifen significantly increased (p < 0.05) the GST specific activity as compared to the drug-treated groups. Glutathione S-transferase acts as a catalyst in conjugation reactions of glutathione with electrophilic substrate and converts it into a more polar form thus leads to the detoxification of xenobiotics by enhancing their excretion (Yuan et al., 2020; Forster et al., 2021).
Cimetidine, rifampicin, dexamethasone and tamoxifen significantly (*p < 0.05) decreased total glutathione level, CAT activity and SOD level; however, significantly (*p < 0.05) increased MDA level in comparison to the control group. C. oppositifolia aqueous and methanolic extract group showed insignificant change in total glutathione level, CAT activity, MDA level and SOD level in comparison to the control group. C. oppositifolia aqueous and methanolic extract co-administered with cimetidine, rifampicin, dexamethasone and tamoxifen significantly increased total glutathione level, CAT activity, and SOD level; however, significantly (*p < 0.05) decreased MDA level in comparison to the drug-treated groups (Li et al., 2019; Alyami et al., 2021).
Conclusions and Recommendations
C. oppositifolia extracts decreased the mRNA expression of phase I drug-metabolizing enzymes; however, increased the glutathione S-transferase activity (phase II drug-metabolizing enzyme). Therefore, caution is needed when C. oppositifolia extracts are to be co-administered with the substrates of CYP1A2, CYP2C9, CYP3A4, and CYP2E1 because extracts may decrease the metabolism of these drugs and chances of toxicity are more. Thus, it is concluded that C. oppositifolia extracts have the potential of pharmacokinetic interactions.
Acknowledgement
The authors would like to thank Punjab University College of Pharmacy, University of the Punjab Lahore, Pakistan for their financial support to conduct this project.
Novelty Statement
This study could be useful to prevent herb-drug interaction. This specially may prove beneficial with Tamoxifen.
Author Contribution
Ammara Abd-Us-Sattar: Performed experiments.
Rukhsana Anwar: Designed and supervised the study.
Shah Jahan: Co-supervised the research.
Afifa Noor: Analyzed the data.
Kanwal Ashiq: Wrote manuscript.
Conflict of interest
The authors have declared no conflict of interest.
References
Ajaib, M., S. Abid, M. Anjum, Q. Noshad, M.F. Siddiqui and M.A. Iqbal. 2011. Phytochemical, antibacterial and antifungal activities of leaves and bark of Colebrookea oppositifolia: An ethnomedicinal plant. Pure App. Biol., 7(1): 138-151. https://doi.org/10.19045/bspab.2018.70017
Alyami, B.A., S. Akhtar, T. Ahmad, A.O. Alqarni, Y.S. Alqahtani, M.H. Mahnashi, S. Qasim, H.M. Irfan, M. Akram, H. Riaz and R. Anwar. 2021. Evaluation of phytochemical, anti-oxidant and cardiac depressant effect of Rumex dentatus by using Langendorff’s isolated heart apparatus. Pak. J. Pharm. Sci., 34: 671-677.
Anwar, R., A.H. Hussin, S. Ismail and S.M. Mansor. 2012. In vitro effect of mitragynine (a major alkaloid of Mitragyna speciosa korth) on aminopyrine metabolism in rat hepatocytes. Int. J. Pharm. Sci. Res., 3(7): 2238.
Anwar, R., R. Sultan and F. Batool. 2018. Ameliorating effect of Berberis lycium root bark extracts against cisplatin-induced nephropathy in rat. Bangladesh J. Pharmacol., 13(3): 248-254. https://doi.org/10.3329/bjp.v13i3.36705
Ashiq, K., K. Hussain, M. Islam, N. Shehzadi, E. Ali and S. Ashiq. 2021. Medicinal plants of Pakistan and their xanthine oxidase inhibition activity to treat gout: A systematic review. Turk. J. Bot., 45(8): 723-738. https://doi.org/10.3906/bot-2109-19
Babos, M.B., M. Heinan, L. Redmond, F. Moiz, J.V. Souza-Peres, V. Samuels and P. Herscu. 2021. Herb drug interactions: Worlds intersect with the patient at the center. Medicines, 8(8): 44. https://doi.org/10.3390/medicines8080044
Baxter, K. and C. Preston. 2010. Stockley’s drug interactions (Vol. 495): Pharmaceutical Press London.
Cascorbi, I., 2012. Drug interactions principles, examples and clinical consequences. Dtsh. Ärztebl. Int.,109(33-34): 546.
Chang, C.C., M.H. Yang, H.M. Wen and J.C. Chern. 2002. Estimation of total flavonoid content in propolis by two complementary colorimetric methods. J. Food Drug. Anal., 10(3): 178-182. https://doi.org/10.38212/2224-6614.2748
Chappell, K.B., S.P. Shah, B.T. Cutshall and C.W. Sands. 2020. Ciprofloxacin-rasagiline drug interaction leading to dopaminergic effects. Nurse Practit., 45(5): 11-12. https://doi.org/10.1097/01.NPR.0000660368.67226.ad
Crewe, H.K., L.M. Notley, R.M. Wunsch, M.S. Lennard and E.M. Gillam. 2002. Metabolism of tamoxifen by recombinant human cytochrome P450 enzymes: formation of the 4-hydroxy, 4′-hydroxy andN-desmethyl metabolites and isomerization oftrans-4-hydroxytamoxifen. Drug. Metab. Disposit., 30(8): 869-874. https://doi.org/10.1124/dmd.30.8.869
Forster, J., J. Duis and M.G. Butler. 2021. Pharmacogenetic testing of cytochrome P450 drug metabolizing enzymes in a case series of patients with prader-willi syndrome. Genes, 12(2): 152. https://doi.org/10.3390/genes12020152
Habig, W.H., M.J. Pabst and W.B. Jakoby. 1974. Glutathione S-transferases: The first enzymatic step in mercapturic acid formation. J. Biol. Chem., 249(22): 7130-7139. https://doi.org/10.1016/S0021-9258(19)42083-8
Hussain, K., Z. Ismail, A. Sadikun and P. Ibrahim. 2008. Analysis of proteins, polysaccharides, glycosaponins contents of Piper sarmentosum Roxb. and anti-TB evaluation for bio-enhancing/interaction effects of leaf extracts with Isoniazid (INH). Nat. Prod. Rad., 7(5): 402-408.
Ijaz, M., M. Fatima, R. Anwar and M. Uroos. 2021. Green synthesis of gold nanoparticles from Manilkara zapota L. extract and the evaluation of its intrinsic in vivo antiarthritic potential. RSC Adv., 11: 27092- 27106. https://doi.org/10.1039/D1RA03186D
Kim, M., S.C. Jee, K.S. Kim, H.S. Kim, K.N. Yu and J.S. Sung. 2021. Quercetin and isorhamnetin attenuate benzo [a] pyrene-induced toxicity by modulating detoxification enzymes through the AhR and NRF2 signaling pathways. Antioxid, 10(5): 787-102. https://doi.org/10.3390/antiox10050787
Latif, A., K. Ashiq, S. Ashiq, E. Ali, I. Anwer and S. Qamar. 2020. Phytochemical analysis and in vitro investigation of anti-inflammatory and xanthine oxidase inhibition potential of root extracts of Bryophyllum pinnatum. J. Anim. Plant Sci., 30(1): 219-228. https://doi.org/10.36899/JAPS.2020.1.0025
Li, A.J., V.K. Pal and K. Kannan. 2021. A review of environmental occurrence, toxicity, biotransformation and biomonitoring of volatile organic compounds. Environ. Chem. Ecotoxicol., 3: 91-116. https://doi.org/10.1016/j.enceco.2021.01.001
Li, Y., Q. Meng, M. Yang, D. Liu, X. Hou, L. Tang, X. Wang, Y. Lyu, X. Chen, K. Liu and A.M. Yu. 2019. Current trends in drug metabolism and pharmacokinetics. Acta Pharm. Sin. B, 9(6): 1113-1144. https://doi.org/10.1016/j.apsb.2019.10.001
Lin, J.H., M. Chiba, I.W. Chen, J.A. Nishime, F.A. deLuna, M. Yamazaki and Y.J. Lin. 1999. Effect of dexamethasone on the intestinal first-pass metabolism of indinavir in rats: Evidence of cytochrome P-450 A and P-glycoprotein induction. Drug. Metab. Disposit., 27(10): 1187-1193.
Lowry, O., N. Rosebrough, A. Farr and R. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Boil. Chem., 193: 265-275. https://doi.org/10.1016/S0021-9258(19)52451-6
Madhavan, V., D.K. Yadav, M. Gurudeva and S. Yoganarasimhan. 2011. Pharmacognostical studies on the leaves of Colebrookea oppositifolia Smith. Asian. J. Trad. Med., 6(4): 134-144.
Magnani, L., E.M. Gaydou and J.C. Hubaud. 2000. Spectrophotometric measurement of antioxidant properties of flavones and flavonols against superoxide anion. Anal. Chim. Acta, 411(1-2): 209-216. https://doi.org/10.1016/S0003-2670(00)00717-0
Martínez, C., C. Albet, J.A. Agúndez, E. Herrero, J.A. Carrillo, M. Márquez, J. Benítez and J.A. Ortiz. 1999. Comparative in vitro and in vivo inhibition of cytochrome P450 CYP1A2, CYP2D6, and CYP3A by H2-receptor antagonists. Clin. Pharmacol. Ther., 65(4): 369-376. https://doi.org/10.1016/S0009-9236(99)70129-3
Mushtaq, A., R. Anwar and M. Ahmad. 2018. Lavandula stoechas (L) a very potent antioxidant attenuates dementia in scopolamine induced memory deficit mice. Front. Pharmacol., 9: 1375. https://doi.org/10.3389/fphar.2018.01375
Mushtaq, A., R. Anwar and M. Ahmad. 2020. Methanolic extract of Pimpinella anisum L. prevents dementia by reducing oxidative stress in neuronal pathways of hypermnesic mice. Pak. J. Zool., 52(5): 1779-1786. https://doi.org/10.17582/journal.pjz/20181002061042
Ohkawa, H., N. Ohishi and K. Yagi. 1979. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem., 95(2): 351-358. https://doi.org/10.1016/0003-2697(79)90738-3
Pleuvry, B.J., 2005. Pharmacodynamic and pharmacokinetic drug interactions. Anaesth. Intensive. Care. Med., 6(4): 129-133. https://doi.org/10.1383/anes.6.4.129.63634
Qayyum, M., K. Ashiq, S. Tanveer, M.A. Bajwa, A. Shah-Rukh and N. Jahangir. 2020. Review on phytochemical evaluation and extraction of Nigella sativa (Kalongi) with pharmacological and traditional applications. Int. J. Biosci., 16(3): 231-241.
Rushmore, T.H. and K.A. Tony. 2002. Pharmacogenomics, regulation and signaling pathways of phase I and II drug metabolizing enzymes. Curr. Drug. Met., 3(5): 481-490. https://doi.org/10.2174/1389200023337171
Sardar, P.R. and S.R. Manik. 2017. GC-MS analysis of aromatic compounds from leaves of Colebrookea oppositifolia Smith. Int. J. Life Sci., 5(2): 241-246.
Sedlak, J. and R.H. Lindsay. 1968. Estimation of total, protein-bound, and nonprotein sulfhydryl groups in tissue with Ellman’s reagent. Anal. Biochem., 25: 192-205. https://doi.org/10.1016/0003-2697(68)90092-4
Sharma, R., Y. Yang, A. Sharma, S. Awasthi and Y.C. Awasthi. 2004. Antioxidant role of glutathione S-transferases: Protection against oxidant toxicity and regulation of stress-mediated apoptosis. Antioxid. Red. Signal., 6(2): 289-300. https://doi.org/10.1089/152308604322899350
Shirsat, R., S. Suradkar and D. Koche. 2014. Preliminary phytochemistry and antimicrobial activity of Salvia plebeia R. Br. and Colebrookea oppositifolia smith. Int. J. Pure. App. Sci. Technol., 20(1): 21.
Sinha, A.K., 1972. Colorimetric assay of catalase. Anal. Biochem., 47(2): 389-394. https://doi.org/10.1016/0003-2697(72)90132-7
Slinkard, K. and V.L. Singleton. 1977. Total phenol analysis: Automation and comparison with manual methods. Am. J. Enol. Vit., 28(1): 49-55.
Tanveer, S.A., A.B. Latif, K.A. Ashiq, M.E. Qayyum and M.A. Bajwa. 2019. A comprehensive review on pharmacological and phytochemical potential of Cassia fistula Linn: A magical herb. Int. J. Biol. Pharm. Allied. Sci., 8(6): 1134-1157. https://doi.org/10.31032/IJBPAS/2019/8.6.4734
Viswanatha, G.L., H. Shylaja, H.Y. Kumar, M.V. Venkataranganna and N.B. Prasad. 2021. Traditional uses, phytochemistry, and ethnopharmacology of Colebrookea oppositifolia Smith: A mini-review. Adv. Trad. Med., 21(2): 209-229. https://doi.org/10.1007/s13596-020-00513-y
Yadav, D.K., 2019. Pharmacognostical, phytochemical and pharmacological profile of Colebrookea oppositifolia Smith. J. Drug. Del. Therap., 9(6-s): 233-237. https://doi.org/10.22270/jddt.v9i6-s.3745
Yemm, E.W. and A. Willis. 1954. The estimation of carbohydrates in plant extracts by anthrone. J. Biochem., 57(3): 508-514. https://doi.org/10.1042/bj0570508
Yuan, X., H. Lu, A. Zhao, Y. Ding, Q. Min and R. Wang. 2020. Transcriptional regulation of CYP3A4 by nuclear receptors in human hepatocytes under hypoxia. Drug Metab. Rev., 52(2): 225-234. https://doi.org/10.1080/03602532.2020.1733004
To share on other social networks, click on any share button. What are these?







